Difference between revisions of "Polyploidy" - New World Encyclopedia
({{Contracted}}) |
m (Robot: Remove contracted tag) |
||
| (9 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | {{ | + | {{Copyedited}}{{Paid}}{{Approved}}{{Images OK}}{{Submitted}} |
| − | '''Polyploidy''' is the condition | + | '''Polyploidy''' is the condition whereby a biological [[cell (biology)|cell]] or [[organism]] has more than two homologous sets of [[chromosome]]s, with each set essentially coding for all the biological traits of the organism. A haploid (n) only has one set of chromosomes. A diploid cell (2n) has two sets of chromosomes. Polyploidy involves three or more times the haploid number of chromosomes. ''Polyploid'' types are termed according to the number of chromosome sets in the [[cell nucleus|nucleus]]: ''triploid'' (three sets; 3n), ''tetraploid'' (four sets; 4n), ''pentaploid'' (five sets; 5n), ''hexaploid'' (six sets; 6n), and so on. |
| − | + | To define this further, homologous chromosomes are those non-identical chromosomes that contain information for the same biological features and contain the same [[gene]]s at the same loci, but possibly different genetic information, called [[allele]]s, at those genes. For example, two chromosomes may have genes encoding eye color, but one may code for brown eyes, the other for blue. Non-homologous chromosomes, representing all the biological features of an organism, form one set, and the number of sets in a cell is called ploidy. In diploid organisms (most [[plant]]s and [[animal]]s), each homologous chromosome is inherited from a different parent. But polyploid organisms have more than two homologous chromosomes. | |
| − | + | Typically, a [[gamete]] or reproductive cell is haploid, while the somatic or body cell of the organism is diploid. That is, a somatic cell has a paired set of chromosomes; the haploid reproductive cell has a single set of unpaired chromosomes, or one half the number of chromosomes of a somatic cell. In diploid organisms, [[sexual reproduction]] involves alternating haploid (n) and diploid (2n) phases, with fusion of haploid cells to produce a diploid organism. (See [[life cycle]].) Some organisms, however, exhibit polyploidy, whereby there are more than two homologous sets of chromosomes. | |
| − | Polyploidy occurs in some [[animal]]s, such as [[goldfish]], [[salmon]], and [[salamander]]s, | + | In addition to being a natural phenomena, [[human being]]s have used polyploidy creatively to create seedless [[banana]]s, hybrids of different species (triticale, a hybrid of [[wheat]] and [[rye]]), and other desirable or more robust plants. |
| + | |||
| + | Note that haploidy is not restricted to sexual reproduction involving [[meiosis]], but may also occur as a normal stage in an organism's life cycle, such as in [[fern]]s and [[fungi]]. In some instances not all the chromosomes are duplicated and the condition is called [[aneuploidy]]. Where an organism is normally [[diploid]], some spontaneous aberrations may occur that are usually caused by a hampered [[cell division]]. | ||
| + | |||
| + | == Polyploidy in animals and plants== | ||
| + | |||
| + | Polyploidy occurs in some [[animal]]s, such as [[goldfish]], [[salmon]], and [[salamander]]s. | ||
| + | |||
| + | However, polyploidy is especially common among [[fern]]s and flowering [[plant]]s, including both wild and cultivated [[species]]. [[Wheat]], for example, after millennia of hybridization and modification by [[human]]s, has strains that are ''diploid'' (two sets of chromosomes); ''tetraploid'' (four sets of chromosomes), with the common name of durum or [[macaroni]] wheat; and ''hexaploid'' (six sets of chromosomes), with the common name of [[bread]] wheat. | ||
| + | |||
| + | Many agriculturally important plants of the genus ''Brassica'' are also tetraploids. This genus, known as cabbages or mustards, includes [[turnip]]s, [[brussels sprouts]], [[cabbage]], [[cauliflower]], [[broccoli]], mustard seed and other important crops. The ''Triangle of U'' is a theory, developed by a Woo Jang-choon, a Korean botanist who was working in [[Japan]], that says the genomes of three ancestral species of Brassica combined to create the three common tetraploid species ''Brassica juncea'' (Indian mustard), ''Brassica napus'' (Rapeseed, rutabaga), and ''Brassica carinata'' (Ethiopian mustard). | ||
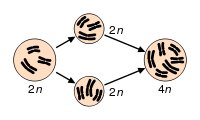
[[Image:Polyploidization.svg|right|frame|300px|Speciation via polyploidy: A [[diploid]] cell undergoes failed [[meiosis]], producing diploid [[gamete]]s, which self-fertilize to produce a tetraploid [[zygote]].]] | [[Image:Polyploidization.svg|right|frame|300px|Speciation via polyploidy: A [[diploid]] cell undergoes failed [[meiosis]], producing diploid [[gamete]]s, which self-fertilize to produce a tetraploid [[zygote]].]] | ||
| − | Examples in animals are more common in the | + | Examples in animals are more common in the lower forms such as [[flatworm]]s, [[leech]]es, and [[brine shrimp]]. Polyploid animals are often sterile, so they often reproduce by [[parthenogenesis]], a form of [[reproduction#Asexual reproduction|asexual reproduction]]. Polyploid salamanders and [[lizard]]s are also quite common and parthenogenetic. While mammalian [[liver]] cells are polyploid, rare instances of polyploid [[mammal]]s are known, but most often result in [[prenatal]] death. |
| − | The only known exception to this rule is an [[Octodontidae|octodontid]] [[rodent]] of [[Argentina]]'s harsh | + | The only known exception to this rule is an [[Octodontidae|octodontid]] [[rodent]] of [[Argentina]]'s harsh desert regions, known as the Red Viscacha-Rat ''(Tympanoctomys barrerae)'', discovered by [http://www.ciencias.uach.cl/index.php Milton Gallardo Narcisi]. This rodent is not a [[rat]], but kin to [[guinea pig]]s and [[chinchilla]]s. Its "new" diploid [2n] number is 102 and so its cells are roughly twice normal size. Its closest living relation is ''Octomys mimax'', the [[Andes|Andean]] Viscacha-Rat of the same family, whose 2n=56. It is surmised that an ''Octomys''-like ancestor produced tetraploid (i.e., 4n=112) offspring that were, by virtue of their doubled chromosomes, reproductively isolated from their parents; but that these likely survived the ordinarily catastrophic effects of polyploidy in mammals by shedding (via chromosomal translocation or some similar mechanism) the "extra" set of sex chromosomes gained at this doubling. |
Polyploidy can be induced in cell culture by some chemicals: the best known is [[colchicine]], which can result in chromosome doubling, though its use may have other less obvious consequences as well. | Polyploidy can be induced in cell culture by some chemicals: the best known is [[colchicine]], which can result in chromosome doubling, though its use may have other less obvious consequences as well. | ||
| − | + | === Polyploid crops === | |
| − | [[ | + | In plant breeding, the induction of polyploids is a common technique to overcome the sterility of a hybrid species. Triticale is the hybrid of wheat ''(Triticum turgidum)'' and [[rye]] ''(Secale cereale)''. It combines sought-after characteristics of the parents, but the initial hybrids are sterile. After polyploidization, the hybrid becomes fertile and can thus be further propagated to become triticale. |
| + | |||
| + | Polyploid plants in general are more robust and sturdy than diploids. In the breeding of crops, those plants that are stronger and tougher are selected. Thus, many crops have unintentionally been bred to a higher level of ploidy: | ||
| + | * Triploid crops: [[banana]], [[apple]], [[ginger]] | ||
| + | * Tetraploid crops: durum or [[macaroni]] [[wheat]], [[maize]], [[cotton]], [[potato]], [[cabbage]], [[Leek (vegetable)|leek]], [[tobacco]], [[peanut]], kinnow, Pelargonium | ||
| + | * Hexaploid crops: [[chrysanthemum]], bread [[wheat]], triticale, [[oat]] | ||
| + | * Octaploid crops: [[strawberry]], [[dahlia]], [[pansies]], [[sugar cane]] | ||
| + | |||
| + | Some crops are found in a variety of ploidy. [[Apple]]s, [[tulip]]s and [[lily|lilies]] are commonly found as both diploid and as triploid. Daylilies ''(Hemerocallis)'' cultivars are available as either diploid or tetraploid. Kinnows can be tetraploid, diploid, or triploid. | ||
| + | |||
| + | In the case of [[banana]]s, while the original bananas contained rather large seeds, triploid (and thus seedless) cultivars have been selected for human consumption. Cultivated bananas are sterile (parthenocarpic), meaning that they do not produce viable seeds. Lacking seeds, another form of propagation is required. These are propagated asexually from offshoots of the plant. | ||
== Polyploidy in humans == | == Polyploidy in humans == | ||
| − | Polyploidy occurs in | + | Polyploidy occurs in [[human]]s in the form of triploidy (69,XXX) and tetraploidy (92,XXXX). |
| − | Triploidy | + | ''Triploidy'' occurs in about two to three percent of all human pregnancies and around 15 percent of miscarriages. The vast majority of triploid conceptions end as [[miscarriage]] and those that do survive to term typically die shortly after birth. In some cases, survival past birth may occur longer if there is mixoploidy, with both a [[diploid]] and a triploid cell population present. |
| − | |||
| − | |||
| − | [[ | + | Triploidy may be the result of either ''diandry'' (the extra haploid set is from the father) or ''digyny'' (the extra haploid set is from the mother). Diandry is almost always caused by the [[fertilization]] of an egg by two [[sperm]] (dispermy). Digyny is most commonly caused by either failure of one [[meiosis|meiotic]] division during oogenesis leading to a diploid oocyte or failure to extrude one polar body from the [[oocyte]]. |
| − | + | Diandry appears to predominate among early miscarriages, while digyny predominates among triploidy that survives into the fetal period. However, among early miscarriages, digyny is also more common in those cases under 8.5 weeks gestational age or those in which an embryo is present. | |
| − | + | There are also two distinct [[phenotype|phenotypes]] in triploid [[placenta|placentas]] and [[fetus|fetuses]] that are dependent on the origin of the extra [[haploid]] set. In digyny, there is typically an asymmetric poorly grown fetus, with marked adrenal hypoplasia (incomplete or arrested development of the [[adrenal gland]]s) and a very small placenta. In diandry, the fetus (when present) is typically normally grown or symmetrically growth restricted, with normal adrenal glands and an abnormally large cystic placenta that is called a partial hydatidiform mole. These parent-of-origin effects reflect the effects of genomic imprinting. | |
| − | + | ||
| − | + | Complete ''tetraploidy'' is more rarely diagnosed than triploidy, but is observed in one to two percent of early miscarriages. However, some tetraploid cells are not uncommonly found in [[chromosome]] analysis at prenatal diagnosis and these are generally considered "harmless." It is not clear whether these tetraploid cells simply tend to arise during ''in vitro'' cell culture or whether they are also present in placental cells ''in vivo''. There are, at any rate, very few clinical reports of fetuses/infants diagnosed with tetraploidy mosaicism. | |
| − | |||
| − | |||
| − | + | Mixoploidy is quite commonly observed in human preimplantation embryos and includes haploid/diploid as well as diploid/tetraploid mixed cell populations. It is unknown whether these embryos fail to implant and are therefore rarely detected in ongoing pregnancies or if there is simply a selective process favoring the diploid cells. | |
== Terminology == | == Terminology == | ||
| Line 44: | Line 60: | ||
=== Autopolyploidy === | === Autopolyploidy === | ||
| − | Autopolyploids are polyploids with | + | ''Autopolyploids'' are polyploids with [[chromosome]]s derived from a single [[species]]. Autopolyploids can arise from a spontaneous, naturally-occurring genome doubling (for example, the [[potato]]). [[Banana]]s and [[apple]]s can be found as triploid autopolyploids. |
=== Allopolyploidy === | === Allopolyploidy === | ||
| − | Allopolyploids are polyploids with chromosomes derived from different species. | + | ''Allopolyploids'' are polyploids with chromosomes derived from different species. [[Triticale]] is an example of an allopolyploid, having six chromosome sets, four from [[wheat]] ''(Triticum turgidum)'' and two from [[rye]] ''(Secale cereale)''. Cabbage is a very interesting example of a fertile allotetraploid crop. ''Amphidiploid'' is another word for an allopolyploid. |
| − | The giant [[tree]] ''[[Sequoia|Sequoia sempervirens]]'' or | + | The giant [[tree]] ''[[Sequoia#Coast Redwood|Sequoia sempervirens]]'' or Coast Redwood has a hexaploid (6n) genome, and is also thought to be autoallopolyploid (AAAABB). |
=== Paleopolyploidy === | === Paleopolyploidy === | ||
| − | Ancient genome duplications probably characterize all life. Duplication events that occurred long ago in the history of various | + | Ancient genome duplications probably characterize all [[life]]. Duplication events that occurred long ago in the history of various lineages can be difficult to detect because of subsequent diploidization (such that a polyploid starts to behave cytogenetically as a diploid over time) as [[mutation]]s and [[gene]] translations gradually make one copy of each chromosome unlike its other copy. |
| − | |||
| − | |||
| − | + | In many cases, these events can be inferred only through comparing sequenced genomes. Examples of unexpected but recently confirmed ancient genome duplications include the baker's [[yeast]] ''(Saccharomyces cerevisiae)'', mustard weed/thale cress ''(Arabidopsis thaliana)'', [[rice]] ''(Oryza sativa)'', and an early ancestor of the [[vertebrate]]s (which includes the [[human]] lineage) and another near the origin of the [[teleost]] [[fish]]es. [[Angiosperm]]s ([[flowering plant]]s) may have paleopolyploidy in their ancestry. All [[eukaryote]]s probably have experienced a polyploidy event at some point in their evolutionary history. | |
| − | |||
| − | |||
==References== | ==References== | ||
| − | + | * Gregory, T. R., and B. K. Mable. 2005. Polyploidy in animals. In T. R. Gregory, ed., ''The Evolution of the Genome''. San Diego: Elsevier. pp. 427-517. ISBN 0123014638. | |
| − | + | * Griffiths, A. J., et al. 2000. ''An Introduction to Genetic Analysis'', 7th ed. New York: W. H. Freeman. ISBN 0-7167-3520-2. | |
| − | + | * Tate, J. A., D. E. Soltis, and P. S. Soltis. 2005. Polyploidy in plants. In T. R. Gregory, ed., ''The Evolution of the Genome''. San Diego: Elsevier. pp. 371-426. ISBN 0123014638. | |
| − | + | * Wolfe, K. H., and D. C. Shields. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. ''Nature'' 387: 708-713. | |
| − | |||
| − | * Gregory, T.R. | ||
| − | |||
| − | * | ||
| − | |||
| − | |||
| − | |||
| − | * Tate, J.A. | ||
| − | |||
| − | |||
| − | |||
| − | * Wolfe, K.H. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | === Further reading === | |
| − | + | * Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant ''Arabidopsis thaliana''. ''Nature'' 408: 796-815. | |
| + | *Eakin, G. S., and R. R. Behringer. 2003. Tetraploid development in the mouse. ''Developmental Dynamics'' 228: 751-766. | ||
| + | * Jaillon, O., et al. 2004. Genome duplication in the teleost fish ''Tetraodon nigroviridis'' reveals the early vertebrate proto-karyotype. ''Nature'' 431: 946-957. | ||
| + | * Paterson, A. H., J. E. Bowers, Y. Van de Peer, and K. Vandepoele. 2005. Ancient duplication of cereal genomes. ''New Phytologist'' 165: 658-661. | ||
| + | *Raes, J., K. Vandepoele, Y. Saeys, C. Simillion, and Y. Van de Peer. 2003. Investigating ancient duplication events in the ''Arabidopsis'' genome. ''Journal of Structural and Functional Genomics'' 3: 117-129. | ||
| + | * Simillion, C., K. Vandepoele, M. Van Montagu, M. Zabeau, and Y. Van de Peer. 2002. The hidden duplication past of ''Arabidopsis thaliana''. ''Proceedings of the National Academy of Science of the USA'' 99: 13627-13632. | ||
| + | * Taylor, J. S., I. Braasch, T. Frickey, A. Meyer, and Y. Van de Peer. 2003. Genome duplication, a trait shared by 22,000 species of ray-finned fish. ''Genome Research'' 13: 382-390. | ||
| + | * Van de Peer, Y., J. S. Taylor, and A. Meyer. 2003. Are all fishes ancient polyploids? ''Journal of Structural and Functional Genomics'' 3: 65-73. | ||
| + | * Van de Peer, Y. 2004. ''Tetraodon'' genome confirms ''Takifugu'' findings: Most fish are ancient polyploids. ''Genome Biology'' 5(12):250. | ||
| + | * Van de Peer, Y., and A. Meyer. 2005. Large-scale gene and ancient genome duplications. In T. R. Gregory, ed., ''The Evolution of the Genome''. San Diego: Elsevier. pp. 329-368. ISBN 0123014638. | ||
| + | * Wolfe, K. H. 2001. Yesterday's polyploids and the mystery of diploidization. ''Nature Reviews Genetics'' 2: 333-341. | ||
[[Category:Life sciences]] | [[Category:Life sciences]] | ||
| − | {{ | + | {{credit5|Polyploidy|105091374|Banana|108171934|Homologous_chromosome|108641008|Triangle_of_U|104253308|Brassica&oldid|105142781}} |
Latest revision as of 00:12, 4 April 2008
Polyploidy is the condition whereby a biological cell or organism has more than two homologous sets of chromosomes, with each set essentially coding for all the biological traits of the organism. A haploid (n) only has one set of chromosomes. A diploid cell (2n) has two sets of chromosomes. Polyploidy involves three or more times the haploid number of chromosomes. Polyploid types are termed according to the number of chromosome sets in the nucleus: triploid (three sets; 3n), tetraploid (four sets; 4n), pentaploid (five sets; 5n), hexaploid (six sets; 6n), and so on.
To define this further, homologous chromosomes are those non-identical chromosomes that contain information for the same biological features and contain the same genes at the same loci, but possibly different genetic information, called alleles, at those genes. For example, two chromosomes may have genes encoding eye color, but one may code for brown eyes, the other for blue. Non-homologous chromosomes, representing all the biological features of an organism, form one set, and the number of sets in a cell is called ploidy. In diploid organisms (most plants and animals), each homologous chromosome is inherited from a different parent. But polyploid organisms have more than two homologous chromosomes.
Typically, a gamete or reproductive cell is haploid, while the somatic or body cell of the organism is diploid. That is, a somatic cell has a paired set of chromosomes; the haploid reproductive cell has a single set of unpaired chromosomes, or one half the number of chromosomes of a somatic cell. In diploid organisms, sexual reproduction involves alternating haploid (n) and diploid (2n) phases, with fusion of haploid cells to produce a diploid organism. (See life cycle.) Some organisms, however, exhibit polyploidy, whereby there are more than two homologous sets of chromosomes.
In addition to being a natural phenomena, human beings have used polyploidy creatively to create seedless bananas, hybrids of different species (triticale, a hybrid of wheat and rye), and other desirable or more robust plants.
Note that haploidy is not restricted to sexual reproduction involving meiosis, but may also occur as a normal stage in an organism's life cycle, such as in ferns and fungi. In some instances not all the chromosomes are duplicated and the condition is called aneuploidy. Where an organism is normally diploid, some spontaneous aberrations may occur that are usually caused by a hampered cell division.
Polyploidy in animals and plants
Polyploidy occurs in some animals, such as goldfish, salmon, and salamanders.
However, polyploidy is especially common among ferns and flowering plants, including both wild and cultivated species. Wheat, for example, after millennia of hybridization and modification by humans, has strains that are diploid (two sets of chromosomes); tetraploid (four sets of chromosomes), with the common name of durum or macaroni wheat; and hexaploid (six sets of chromosomes), with the common name of bread wheat.
Many agriculturally important plants of the genus Brassica are also tetraploids. This genus, known as cabbages or mustards, includes turnips, brussels sprouts, cabbage, cauliflower, broccoli, mustard seed and other important crops. The Triangle of U is a theory, developed by a Woo Jang-choon, a Korean botanist who was working in Japan, that says the genomes of three ancestral species of Brassica combined to create the three common tetraploid species Brassica juncea (Indian mustard), Brassica napus (Rapeseed, rutabaga), and Brassica carinata (Ethiopian mustard).
Examples in animals are more common in the lower forms such as flatworms, leeches, and brine shrimp. Polyploid animals are often sterile, so they often reproduce by parthenogenesis, a form of asexual reproduction. Polyploid salamanders and lizards are also quite common and parthenogenetic. While mammalian liver cells are polyploid, rare instances of polyploid mammals are known, but most often result in prenatal death.
The only known exception to this rule is an octodontid rodent of Argentina's harsh desert regions, known as the Red Viscacha-Rat (Tympanoctomys barrerae), discovered by Milton Gallardo Narcisi. This rodent is not a rat, but kin to guinea pigs and chinchillas. Its "new" diploid [2n] number is 102 and so its cells are roughly twice normal size. Its closest living relation is Octomys mimax, the Andean Viscacha-Rat of the same family, whose 2n=56. It is surmised that an Octomys-like ancestor produced tetraploid (i.e., 4n=112) offspring that were, by virtue of their doubled chromosomes, reproductively isolated from their parents; but that these likely survived the ordinarily catastrophic effects of polyploidy in mammals by shedding (via chromosomal translocation or some similar mechanism) the "extra" set of sex chromosomes gained at this doubling.
Polyploidy can be induced in cell culture by some chemicals: the best known is colchicine, which can result in chromosome doubling, though its use may have other less obvious consequences as well.
Polyploid crops
In plant breeding, the induction of polyploids is a common technique to overcome the sterility of a hybrid species. Triticale is the hybrid of wheat (Triticum turgidum) and rye (Secale cereale). It combines sought-after characteristics of the parents, but the initial hybrids are sterile. After polyploidization, the hybrid becomes fertile and can thus be further propagated to become triticale.
Polyploid plants in general are more robust and sturdy than diploids. In the breeding of crops, those plants that are stronger and tougher are selected. Thus, many crops have unintentionally been bred to a higher level of ploidy:
- Triploid crops: banana, apple, ginger
- Tetraploid crops: durum or macaroni wheat, maize, cotton, potato, cabbage, leek, tobacco, peanut, kinnow, Pelargonium
- Hexaploid crops: chrysanthemum, bread wheat, triticale, oat
- Octaploid crops: strawberry, dahlia, pansies, sugar cane
Some crops are found in a variety of ploidy. Apples, tulips and lilies are commonly found as both diploid and as triploid. Daylilies (Hemerocallis) cultivars are available as either diploid or tetraploid. Kinnows can be tetraploid, diploid, or triploid.
In the case of bananas, while the original bananas contained rather large seeds, triploid (and thus seedless) cultivars have been selected for human consumption. Cultivated bananas are sterile (parthenocarpic), meaning that they do not produce viable seeds. Lacking seeds, another form of propagation is required. These are propagated asexually from offshoots of the plant.
Polyploidy in humans
Polyploidy occurs in humans in the form of triploidy (69,XXX) and tetraploidy (92,XXXX).
Triploidy occurs in about two to three percent of all human pregnancies and around 15 percent of miscarriages. The vast majority of triploid conceptions end as miscarriage and those that do survive to term typically die shortly after birth. In some cases, survival past birth may occur longer if there is mixoploidy, with both a diploid and a triploid cell population present.
Triploidy may be the result of either diandry (the extra haploid set is from the father) or digyny (the extra haploid set is from the mother). Diandry is almost always caused by the fertilization of an egg by two sperm (dispermy). Digyny is most commonly caused by either failure of one meiotic division during oogenesis leading to a diploid oocyte or failure to extrude one polar body from the oocyte.
Diandry appears to predominate among early miscarriages, while digyny predominates among triploidy that survives into the fetal period. However, among early miscarriages, digyny is also more common in those cases under 8.5 weeks gestational age or those in which an embryo is present.
There are also two distinct phenotypes in triploid placentas and fetuses that are dependent on the origin of the extra haploid set. In digyny, there is typically an asymmetric poorly grown fetus, with marked adrenal hypoplasia (incomplete or arrested development of the adrenal glands) and a very small placenta. In diandry, the fetus (when present) is typically normally grown or symmetrically growth restricted, with normal adrenal glands and an abnormally large cystic placenta that is called a partial hydatidiform mole. These parent-of-origin effects reflect the effects of genomic imprinting.
Complete tetraploidy is more rarely diagnosed than triploidy, but is observed in one to two percent of early miscarriages. However, some tetraploid cells are not uncommonly found in chromosome analysis at prenatal diagnosis and these are generally considered "harmless." It is not clear whether these tetraploid cells simply tend to arise during in vitro cell culture or whether they are also present in placental cells in vivo. There are, at any rate, very few clinical reports of fetuses/infants diagnosed with tetraploidy mosaicism.
Mixoploidy is quite commonly observed in human preimplantation embryos and includes haploid/diploid as well as diploid/tetraploid mixed cell populations. It is unknown whether these embryos fail to implant and are therefore rarely detected in ongoing pregnancies or if there is simply a selective process favoring the diploid cells.
Terminology
Autopolyploidy
Autopolyploids are polyploids with chromosomes derived from a single species. Autopolyploids can arise from a spontaneous, naturally-occurring genome doubling (for example, the potato). Bananas and apples can be found as triploid autopolyploids.
Allopolyploidy
Allopolyploids are polyploids with chromosomes derived from different species. Triticale is an example of an allopolyploid, having six chromosome sets, four from wheat (Triticum turgidum) and two from rye (Secale cereale). Cabbage is a very interesting example of a fertile allotetraploid crop. Amphidiploid is another word for an allopolyploid.
The giant tree Sequoia sempervirens or Coast Redwood has a hexaploid (6n) genome, and is also thought to be autoallopolyploid (AAAABB).
Paleopolyploidy
Ancient genome duplications probably characterize all life. Duplication events that occurred long ago in the history of various lineages can be difficult to detect because of subsequent diploidization (such that a polyploid starts to behave cytogenetically as a diploid over time) as mutations and gene translations gradually make one copy of each chromosome unlike its other copy.
In many cases, these events can be inferred only through comparing sequenced genomes. Examples of unexpected but recently confirmed ancient genome duplications include the baker's yeast (Saccharomyces cerevisiae), mustard weed/thale cress (Arabidopsis thaliana), rice (Oryza sativa), and an early ancestor of the vertebrates (which includes the human lineage) and another near the origin of the teleost fishes. Angiosperms (flowering plants) may have paleopolyploidy in their ancestry. All eukaryotes probably have experienced a polyploidy event at some point in their evolutionary history.
ReferencesISBN links support NWE through referral fees
- Gregory, T. R., and B. K. Mable. 2005. Polyploidy in animals. In T. R. Gregory, ed., The Evolution of the Genome. San Diego: Elsevier. pp. 427-517. ISBN 0123014638.
- Griffiths, A. J., et al. 2000. An Introduction to Genetic Analysis, 7th ed. New York: W. H. Freeman. ISBN 0-7167-3520-2.
- Tate, J. A., D. E. Soltis, and P. S. Soltis. 2005. Polyploidy in plants. In T. R. Gregory, ed., The Evolution of the Genome. San Diego: Elsevier. pp. 371-426. ISBN 0123014638.
- Wolfe, K. H., and D. C. Shields. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708-713.
Further reading
- Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815.
- Eakin, G. S., and R. R. Behringer. 2003. Tetraploid development in the mouse. Developmental Dynamics 228: 751-766.
- Jaillon, O., et al. 2004. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431: 946-957.
- Paterson, A. H., J. E. Bowers, Y. Van de Peer, and K. Vandepoele. 2005. Ancient duplication of cereal genomes. New Phytologist 165: 658-661.
- Raes, J., K. Vandepoele, Y. Saeys, C. Simillion, and Y. Van de Peer. 2003. Investigating ancient duplication events in the Arabidopsis genome. Journal of Structural and Functional Genomics 3: 117-129.
- Simillion, C., K. Vandepoele, M. Van Montagu, M. Zabeau, and Y. Van de Peer. 2002. The hidden duplication past of Arabidopsis thaliana. Proceedings of the National Academy of Science of the USA 99: 13627-13632.
- Taylor, J. S., I. Braasch, T. Frickey, A. Meyer, and Y. Van de Peer. 2003. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Research 13: 382-390.
- Van de Peer, Y., J. S. Taylor, and A. Meyer. 2003. Are all fishes ancient polyploids? Journal of Structural and Functional Genomics 3: 65-73.
- Van de Peer, Y. 2004. Tetraodon genome confirms Takifugu findings: Most fish are ancient polyploids. Genome Biology 5(12):250.
- Van de Peer, Y., and A. Meyer. 2005. Large-scale gene and ancient genome duplications. In T. R. Gregory, ed., The Evolution of the Genome. San Diego: Elsevier. pp. 329-368. ISBN 0123014638.
- Wolfe, K. H. 2001. Yesterday's polyploids and the mystery of diploidization. Nature Reviews Genetics 2: 333-341.
Credits
New World Encyclopedia writers and editors rewrote and completed the Wikipedia article in accordance with New World Encyclopedia standards. This article abides by terms of the Creative Commons CC-by-sa 3.0 License (CC-by-sa), which may be used and disseminated with proper attribution. Credit is due under the terms of this license that can reference both the New World Encyclopedia contributors and the selfless volunteer contributors of the Wikimedia Foundation. To cite this article click here for a list of acceptable citing formats.The history of earlier contributions by wikipedians is accessible to researchers here:
- Polyploidy history
- Banana history
- Homologous_chromosome history
- Triangle_of_U history
- Brassica&oldid history
The history of this article since it was imported to New World Encyclopedia:
Note: Some restrictions may apply to use of individual images which are separately licensed.