Difference between revisions of "Nucleotide" - New World Encyclopedia
| Line 10: | Line 10: | ||
==Chemical structure and nomenclature== | ==Chemical structure and nomenclature== | ||
| − | + | The nitrogen-containing base in nucleotides (also called the ''nucleobase'') is typically a derivative of purine or pyrimidine, which are [[heterocylic]] compounds, or organic compounds that contain a ring structure that contains atoms in addition to carbon, such as sulfur, oxygen or nitrogen. The most common bases in nucleotides are: | |
| + | *The purines [[adenine]] and [[guanine]]; and | ||
| + | *The pyrimidines [[cytosime]], [[thymine]], and [[uracil]] | ||
| − | The | + | The sugar component is either deoxyribose or ribose. (“Deoxy” simply indicates that the sugar lacks an oxygen atom present in ribose, the parent compound.) Depending on their base sugar, nucleotides are therefore known as “deoxyribonucleotides” or “ribonucleotides.” The nucleic acid DNA (which stands for ''deoxyribonucleic acid'') is built of nucleotides with a deoxyribose sugar, whereas RNA (or ''ribonucleic acid'') contains nucleotides composed of ribose sugars. |
| − | |||
| − | |||
| − | |||
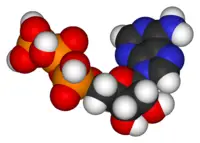
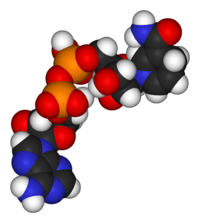
| − | + | [[Image:Nucleotides.png|400px|left|frame|Nucleotides consist of a pentose sugar (brown), one or more phosphate groups (red), and a heterocyclic base (blue). The structures of some of the common bases, which are derivatives of either purine or pyrimidine, are pictured at right.]] | |
| − | Nucleosides resemble the structure of nucleotides (i.e., they contain a purine or | + | Nucleotide names are abbreviated into standard three- or four-letter codes that indicate their structural components: |
| − | + | *The first letter is lower case and indicates whether the nucleotide in question is a deoxyribonucleotide (denoted by a d) or a ribonucleotide (no letter). | |
| − | + | *The second letter indicates the nucleoside corresponding to the base. ''Nucleosides'' resemble the structure of nucleotides (i.e., they contain a purine or pyrimidine base bonded to a sugar) but lack the phosphate group (Stryer 76) A nucleotide can also be defined as the phosphate ester of a nucleotide. In chemistry, esters are organic compounds in which an organic group (symbolized by R' in this article) replaces a hydrogen atom (or more than one) in an oxygen acid. The abbreviations are as follows: | |
| − | |||
| − | |||
: G: [[Guanine]] | : G: [[Guanine]] | ||
: A: [[Adenine]] | : A: [[Adenine]] | ||
: T: [[Thymine]] | : T: [[Thymine]] | ||
: C: [[Cytosine]] | : C: [[Cytosine]] | ||
| − | : U: [[Uracil]] | + | : U: [[Uracil]] (which is not present in DNA, but takes the place of thymine in RNA) |
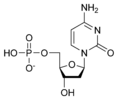
| − | The third and fourth letters indicate the length of the attached phosphate chain (Mono-, Di-, Tri-) and the presence of a phosphate (P). | + | *The third and fourth letters indicate the length of the attached phosphate chain (Mono-, Di-, Tri-) and the presence of a phosphate (P). |
| + | |||
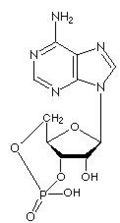
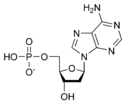
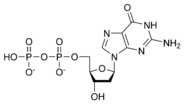
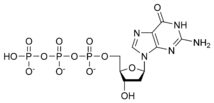
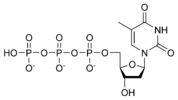
| + | [[Image:dATP_chemical_structure.png|186px|right|frame|Chemical structure of deoxyadenosine triphosphate]] | ||
| − | Thus, for example, deoxy- | + | Thus, for example, deoxy-adenosine-triphosphate (pictured at right), one of the activated precursors in the synthesis of DNA, is abbreviated as dATP. |
==Nucleotides encode genetic information== | ==Nucleotides encode genetic information== | ||
Revision as of 04:58, 17 August 2006
A nucleotide is a chemical compound with three components: a nitrogen-containing base, a pentose (five-carbon) sugar, and one or more phosphate groups. Although best known as the structural units of the nucleic acids DNA and RNA, which encode genetic information in organisms, nucleotides participate in nearly all biochemical processes.
Nucleotides' role in metabolism:
- ATP, an adenine nucleotide, is a universal energy currency of energy in biological systems.
- Adenine nucleotides are components of three major coenzymes, NAD plus, FAD, and CoA
- Nucleotides also serve as regulators of metabolism. Cyclic AMP is a ubiquitous mediator of the action of many hormones. Covalent modifications introduced by ATP alter the activities of many enzymes (e.g., phosphorylation of glycogen synthase)
perhaps explain reason for ubiquity of ribonucleotides in particular—evolutionarily, RNA is believed to have come before DNA and proteins. (Stryer 115 and ch. 17) (also see rna-world hypothesis in wikipedia)
Chemical structure and nomenclature
The nitrogen-containing base in nucleotides (also called the nucleobase) is typically a derivative of purine or pyrimidine, which are heterocylic compounds, or organic compounds that contain a ring structure that contains atoms in addition to carbon, such as sulfur, oxygen or nitrogen. The most common bases in nucleotides are:
The sugar component is either deoxyribose or ribose. (“Deoxy” simply indicates that the sugar lacks an oxygen atom present in ribose, the parent compound.) Depending on their base sugar, nucleotides are therefore known as “deoxyribonucleotides” or “ribonucleotides.” The nucleic acid DNA (which stands for deoxyribonucleic acid) is built of nucleotides with a deoxyribose sugar, whereas RNA (or ribonucleic acid) contains nucleotides composed of ribose sugars.
Nucleotide names are abbreviated into standard three- or four-letter codes that indicate their structural components:
- The first letter is lower case and indicates whether the nucleotide in question is a deoxyribonucleotide (denoted by a d) or a ribonucleotide (no letter).
- The second letter indicates the nucleoside corresponding to the base. Nucleosides resemble the structure of nucleotides (i.e., they contain a purine or pyrimidine base bonded to a sugar) but lack the phosphate group (Stryer 76) A nucleotide can also be defined as the phosphate ester of a nucleotide. In chemistry, esters are organic compounds in which an organic group (symbolized by R' in this article) replaces a hydrogen atom (or more than one) in an oxygen acid. The abbreviations are as follows:
- G: Guanine
- A: Adenine
- T: Thymine
- C: Cytosine
- U: Uracil (which is not present in DNA, but takes the place of thymine in RNA)
- The third and fourth letters indicate the length of the attached phosphate chain (Mono-, Di-, Tri-) and the presence of a phosphate (P).
Thus, for example, deoxy-adenosine-triphosphate (pictured at right), one of the activated precursors in the synthesis of DNA, is abbreviated as dATP.
Nucleotides encode genetic information
DNA is a polymer of deoxyribonucleotide units. Polymer is a term used to describe molecules consisting of structural units and a large number of repeating units connected by covalent chemical bonds.
In DNA, bases carry genetic information while sugar and phosphate groups provide a structural role (as the base is the variable part).
Structure of DNA: two helical polynucleotide chains are coiled around a common axis. The chains run in opposite directions. The bases are on the inside of the helix, while the sugar-phosphate backbone is on the outside. The two chains are held together by hydrogen bonds between pairs of bases. Adenine is always paired with thymine; guanine with cytosine (i.e., a purine to a pyrimidine).
Two differences of RNA, which can serve as genetic material in viruses: (1) sugar units are riboses rather than deoxyriboses and (2) one of the four major bases is uracil (U) instead of thymine (T); can be single or double-stranded
RNA molecules can contain as few as 75 nucleotides to more than 5000 nucleotides.
The genetic code is the relation between the sequence of bases in DNA (or its RNA transcript) and the sequence of amino acids in proteins. Amino acids are coded by groups of three bases (called codons) starting from a fixed point.
Although sometimes called "the molecule of heredity", DNA macromolecules as people typically think of them are not single molecules. Rather, they are pairs of molecules, which entwine like vines to form a double helix (see the illustration at the right).
Each vine-like molecule is a strand of DNA: a chemically linked chain of nucleotides, each of which consists of a sugar (deoxyribose), a phosphate and one of five kinds of nucleobases ("bases"). Because DNA strands are composed of these nucleotide subunits, they are polymers.
The diversity of the bases means that there are five kinds of nucleotides, which are commonly referred to by the identity of their bases. These are adenine (A), thymine (T), uracil (U), cytosine (C), and guanine (G). U is rarely found in DNA except as a result of chemical degradation of C, but in some viruses, notably PBS1 phage DNA, U completely replaces the usual T in its DNA. Similarly, RNA usually contains U in place of T, but in certain RNAs such as transfer RNA, T is always found in some positions. Thus, the only true difference between DNA and RNA is the sugar, 2-deoxyribose in DNA and ribose in RNA.
In a DNA double helix, two polynucleotide strands can associate through the hydrophobic effect and pi stacking. Specificity of which strands stay associated is determined by complementary pairing. Each base forms hydrogen bonds readily to only one other, A to T forming two hydrogen bonds and C to G forming three hydrogen bonds. The GC content and length of each DNA molcule dictates the strength of the association; the more complementary bases exist, the stronger and longer-lasting the association characterised by the temperature required to break the hydrogen bonding, its Tm value.
The cell's machinery is capable of melting or disassociating a DNA double helix, and using each DNA strand as a template for synthesizing a new strand which is nearly identical to the previous strand. Errors that occur in the synthesis are known as mutations. The process known as PCR (polymerase chain reaction) mimics this process in vitro in a nonliving system.
Because pairing causes the nucleotide bases to face the helical axis, the sugar and phosphate groups of the nucleotides run along the outside; the two chains they form are sometimes called the "backbones" of the helix. In fact, it is chemical bonds between the phosphates and the sugars that link one nucleotide to the next in the DNA strand.
Within a gene, the sequence of nucleotides along a DNA strand defines a messenger RNA sequence which then defines a protein, that an organism is liable to manufacture or "express" at one or several points in its life using the information of the sequence. The relationship between the nucleotide sequence and the amino-acid sequence of the protein is determined by simple cellular rules of translation, known collectively as the genetic code. The genetic code consists of three-letter 'words' (termed a codon) formed from a sequence of three nucleotides (e.g. ACT, CAG, TTT). These codons can then be translated with messenger RNA and then transfer RNA, with a codon corresponding to a particular amino acid. There are 64 possible codons (4 bases in 3 places ) that encode 20 amino acids. Most amino acids, therefore, have more than one possible codon. There are also three 'stop' or 'nonsense' codons signifying the end of the coding region, namely the UAA, UGA and UAG codons.
Messenger RNA is RNA that carries information from DNA to the ribosome sites of protein synthesis in the cell. Once mRNA has been transcribed from DNA, it is exported from the nucleus into the cytoplasm (in eukaryotes mRNA is "processed" before being exported), where it is bound to ribosomes and translated into protein. After a certain amount of time the message degrades into its component nucleotides, usually with the assistance of RNA polymerases.
Transfer RNA is a small RNA chain of about 74-93 nucleotides that transfers a specific amino acid to a growing polypeptide chain at the ribosomal site of protein synthesis during translation. It has sites for amino-acid attachment and an anticodon region for codon recognition that binds to a specific sequence on the messenger RNA chain through hydrogen bonding. It is a type of non-coding RNA.
Ribosomal RNA (rRNA) is a component of the ribosomes, the protein synthetic factories in the cell. Eukaryotic ribosomes contain four different rRNA molecules: 18S, 5.8S, 28S, and 5S rRNA. Three of the rRNA molecules are synthesized in the nucleolus, and one is synthesized elsewhere. rRNA molecules are extremely abundant. They make up at least 80% of the RNA molecules found in a typical eukaryotic cell.
Nucleotides function in cell metabolism
ATP is the universal energy currency of the cell
multifunctional nucleotide primarily known in biochemistry as the "molecular currency" of intracellular energy transfer. In this role ATP transports chemical energy within cells. It is produced as an energy source during the processes of photosynthesis and cellular respiration. The structure of this molecule consists of a purine base (adenine) attached to the 1' carbon atom of a pentose (ribose). Three phosphate groups are attached at the 5' carbon atom of the pentose. ATP is also one of four monomers (nucleotides) required for the synthesis of ribonucleic acids.
ATP energy is released when hydrolysis of the phosphate-phosphate bonds is carried out. This energy can be used by a variety of enzymes, motor proteins, and transport proteins to carry out the work of the cell. Also, the hydrolysis yields free inorganic Pi and ADP, which can be broken down further to another Pi and AMP. ATP can also be broken down to AMP directly, with the formation of PPi. This last reaction has the advantage of being an effectively irreversible process in aqueous solution.
A few examples of the use of ATP include the active transport of molecules across cell membranes, the synthesis of macromolecules (Eg. proteins), muscle contractions, endocytosis, and exocytosis.
The total quantity of ATP in the human body is about 0.1 mole. The energy used by human cells requires the hydrolysis of 200 to 300 moles of ATP daily. This means that each ATP molecule is recycled 2000 to 3000 times during a single day. ATP cannot be stored, hence its consumption must closely follow its synthesis.
- Note that ATP isn't usually actually synthethised from scratch, but simply recharged from ADP (Adenosine Diphosphate). The amount of ATP + ADP in your body remains roughly the same, it just keeps cycling.
ATP can be produced by various cellular processes: Under aerobic conditions, the majority of the synthesis occurs in mitochondria during oxidative phosphorylation and is catalyzed by ATP synthase and, to a lesser degree, under anaerobic conditions by fermentation.
also mention other nucleotides that have high-energy phosphate bonds
Several nucleotides function as coenzymes
Nicotinamide adenine dinucleotide (NAD+) and nicotinamide adenine dinucleotide phosphate (NADP) are two important cofactors found in cells. NADH is the reduced form of NAD+, and NAD+ is the oxidized form of NADH. It forms NADP with the addition of a phosphate group to the 2' position of the adenosyl nucleotide through an ester linkage.
NAD is used extensively in glycolysis and the citric acid cycle of cellular respiration. The reducing potential stored in NADH can be converted to ATP through the electron transport chain or used for anabolic metabolism. ATP "energy" is necessary for an organism to live. Green plants obtain ATP through photosynthesis, while other organisms obtain it by cellular respiration.
NADP is used in anabolic reactions, such as fat acid and nucleic acid synthesis, that require NADPH as a reducing agent. In chloroplasts, NADP is an oxidising agent important in the preliminary reactions of photosynthesis. The NADPH produced by photosynthesis is then used as reducing power for the biosynthetic reactions in the Calvin cycle of photosynthesis.
sentence or two on fad
Coenzyme A (CoA, CoASH, or HSCoA) is a coenzyme, notable for its role in the synthesis and oxidization of fatty acids, and the oxidation of pyruvate in the citric acid cycle. It is adapted from β-mercaptoethylamine, panthothenate and adenosine triphosphate.
The main function of coenzyme A is to carry acyl groups (such as the acetyl group) or thioesters. A molecule of coenzyme A carrying an acetyl group is also referred to as acetyl-CoA. It is sometimes referred to as 'CoASH' or 'HSCoA' because when it is not attached to a molecule such as an acetyl group, it is attached to a thiol group, -SH.
Acetyl-CoA is an important molecule itself. It is the precursor to HMG CoA, which is a vital component in cholesterol and ketone synthesis. Furthermore, it contributes an acetyl group to choline to produce acetylcholine, in a reaction catalysed by choline acetyltransferase. Its main task is conveying the carbon atoms within the acetyl group to the citric acid cycle to be oxidized for energy production.
Nucleotides also play a role in regulation and signaling
Cyclic adenosine monophosphate (cAMP, cyclic AMP or 3'-5'-cyclic adenosine monophosphate) is a molecule that is important in many biological processes; it is derived from adenosine triphosphate (ATP). cAMP is a second messenger, used for intracellular signal transduction, such as transferring the effects of hormones like glucagon and adrenaline, which cannot get through the cell membrane. Its main purpose is the activation of protein kinases; it is also used to regulate the passage of Ca2+ through ion channels.
cAMP is synthesised from ATP by adenylate cyclase. Adenylate cyclase is located at the cell membranes. It is activated by the hormones glucagon and adrenaline and by G protein. Liver adenylate cyclase responds more strongly to glucagon, and muscle adenylate cyclase responds more strongly to adrenaline.
cAMP decomposition into AMP is catalyzed by the enzyme phosphodiesterase. This enzyme is inhibited by high concentrations of caffeine, so it is possible that the stimulatory effect of this drug is the result of the raised cAMP levels that it causes (However it seems the concentrations required for caffeine to be effective are very high and a more likely explanation for the drug's effects involve the adenosine molecule).
Cyclic AMP is involved in some protein kinases. For example, PKA (protein kinase A, also known as cAMP-dependent protein kinase) is normally inactive as a tetrameric holoenzyme, consisting of 2 catalytic and 2 regulatory units (C2R2), with the regulatory units blocking the catalytic centers of the catalytic units.
Cyclic AMP binds to specific locations on the regulatory units of the protein kinase, and causes dissociation between the regulatory and catalytic subunits, thus activating the catalytic units and enabling them to phosphorylate substrate proteins.
cAMP controls many biological processes, including glycogen decomposition into glucose (glycogenolysis), and lipolysis.
In bacteria, the level of cAMP varies depending on the medium used for growth. In particular, cAMP is low when glucose is the carbon source. This occurs through inhibition of the cAMP-producing enzyme, adenylate cyclase, as a side effect of glucose transport into the cell. The transcription factor CRP (or CAP) forms a complex with cAMP and thereby is activated to bind to DNA. CRP-cAMP increases expression of a large number of genes, including some encoding enzymes that can supply energy independent of glucose.
Chemical structures
Nucleotides
 Adenosine monophosphate AMP |
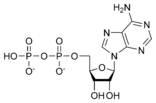 Adenosine diphosphate ADP |
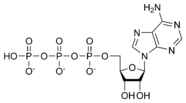 Adenosine triphosphate ATP |
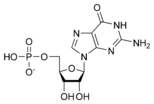 Guanosine monophosphate GMP |
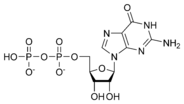 Guanosine diphosphate GDP |
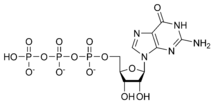 Guanosine triphosphate GTP |
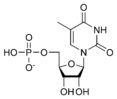 Thymidine monophosphate TMP |
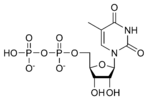 Thymidine diphosphate TDP |
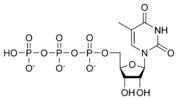 Thymidine triphosphate TTP |
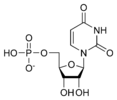 Uridine monophosphate UMP |
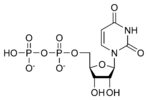 Uridine diphosphate UDP |
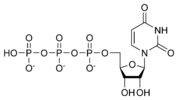 Uridine triphosphate UTP |
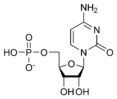 Cytidine monophosphate CMP |
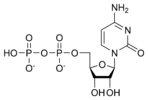 Cytidine diphosphate CDP |
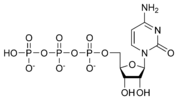 Cytidine triphosphate CTP |
Deoxynucleotides
| Code | Equivalence | Complement |
|---|---|---|
| A | A | T or U |
| C | C | G |
| G | G | C |
| T or U | T | A |
| M | A or C | K |
| R | A or G | Y |
| W | A or T | W |
| S | C or G | S |
| Y | C or T | R |
| K | G or T | M |
| V | A or C or G | B |
| H | A or C or T | D |
| D | A or G or T | H |
| B | C or G or T | V |
| X or N | A or C or G or T | X |
ReferencesISBN links support NWE through referral fees
- Stryer, Lubert. 1995. Biochemistry, 4th edition. New York, NY: W.H. Freeman.
External links
- Abbreviations and Symbols for Nucleic Acids, Polynucleotides and their Constituents (IUPAC)
- Provisional Recommendations 2004 (IUPAC)
| Nucleic acids edit |
|---|
| Nucleobases: Adenine - Thymine - Uracil - Guanine - Cytosine - Purine - Pyrimidine |
| Nucleosides: Adenosine - Uridine - Guanosine - Cytidine - Deoxyadenosine - Thymidine - Deoxyguanosine - Deoxycytidine |
| Nucleotides: AMP - UMP - GMP - CMP - ADP - UDP - GDP - CDP - ATP - UTP - GTP - CTP - cAMP - cGMP |
| Deoxynucleotides: dAMP - dTMP - dUMP - dGMP - dCMP - dADP - dTDP - dUDP - dGDP - dCDP - dATP - dTTP - dUTP - dGTP - dCTP |
| Nucleic acids: DNA - RNA - LNA - PNA - mRNA - ncRNA - miRNA - rRNA - siRNA - tRNA - mtDNA - Oligonucleotide |
Credits
New World Encyclopedia writers and editors rewrote and completed the Wikipedia article in accordance with New World Encyclopedia standards. This article abides by terms of the Creative Commons CC-by-sa 3.0 License (CC-by-sa), which may be used and disseminated with proper attribution. Credit is due under the terms of this license that can reference both the New World Encyclopedia contributors and the selfless volunteer contributors of the Wikimedia Foundation. To cite this article click here for a list of acceptable citing formats.The history of earlier contributions by wikipedians is accessible to researchers here:
The history of this article since it was imported to New World Encyclopedia:
Note: Some restrictions may apply to use of individual images which are separately licensed.