RNA
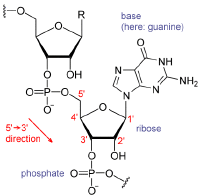
Ribonucleic acid or RNA is a polymer or chain of nucleotide units, each comprising a nitrogenous base (adenine, cytosine, guanine, or uracil), a five-carbon sugar (ribose), and a phosphate group. The sugar and phosphate groups form the polymer's backbone, while the nitrogenous bases extending from the backbone provide RNA's distinctive properties.
In living cells, RNA in different configurations fulfills several important roles in the process of translating genetic information from deoxyribonucleic acid (DNA) into proteins. One type of RNA (messenger(m) RNA) acts as a messenger between DNA and the protein synthesis complexes known as ribosomes; a second type (ribosomal(r) RNA) forms vital portions of the structure of ribosomes; a third type (transfer(t) RNA) is an essential guide to deliver the appropriate protein building blocks, amino acids, to the ribosome; and other types of RNA, microRNAs (miRNAs) play a role in regulating gene expression, while small nuclear(sn) RNA helps with assuring that mRNA contains no nucleotide units that would lead to formation of a faulty protein. RNA also serves as a genetic blueprint for certain viruses, and some RNA molecules (called ribozymes) are also involved in the catalysis of biochemical reactions.
RNA is very similar to DNA, but differs in a few important structural details. RNA is usually single stranded, while DNA naturally seeks its stable form as a double stranded molecule. RNA nucleotides contain ribose while DNA nucleotides contain the closely related sugar deoxyribose. Furthermore, RNA uses the nucleotide uracil in its composition, instead of the thymine that is present in DNA. RNA is transcribed from DNA by enzymes called RNA polymerases and is generally further processed by other enzymes, some of them guided by non-coding RNAs.
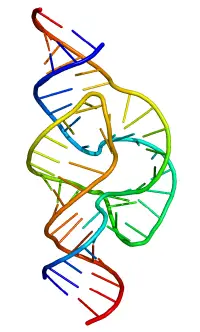
Single-stranded RNA is similar to the protein polymer in its natural propensity to fold back and double up with itself in complex ways assuming a variety of biologically useful configurations.
The connectedness of living organisms can be seen in the ubiquitousness of RNA in living cells and in viruses throughout nature, and in the universal role of RNA in protein synthesis.
Chemical and stereochemical structure
RNA is a nucleic acid, a complex, high-molecular-weight macromolecule composed of nucleotide chains whose sequence of bases conveys genetic information.
A nucleotide is a chemical compound comprising three components: a nitrogen-containing base, a pentose (five-carbon) sugar, and one or more phosphate groups. The nitrogen-containing base of a nucleotide (also called the nucleobase) is typically a derivative of either purine or pyrimidine. The most common nucleotide bases are the purines adenine and guanine and the pyrimidines cytosine and thymine (or uracil in RNA).
Nucleic acids are polymers of repeating units (called monomers). Specifically, they often comprise long chains of nucleotide monomers connected by covalent chemical bonds. RNA molecules may comprise as few as 75 nucleotides or more than 5,000 nucleotides, while a DNA molecule may comprise more than 1,000,000 nucleotide units.
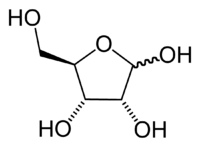
In RNA, the sugar component, ribose is a water-soluable, pentose sugar (monosaccharide with five carbon atoms). Ribose has the chemical formula C5H10O5.
Ribose is an aldopentose, which means a pentose sugar with an aldehyde functional group in position one. An aldehyde group comprises a carbon atom bonded to a hydrogen atom and double-bonded to an oxygen atom (chemical formula O=CH-). Ribose forms a five-member ring with four carbon atoms and one oxygen. Hydroxyl (-OH) groups are attached to three of the carbons. The fourth carbon in the ring (one of the carbon atoms adjacent to the oxygen) has attached to it the fifth carbon atom and a hydroxyl group.
The RNA polymer features a ribose and phosphate backbone with one of four different nucleotide basesâadenine, guanine, cytosine, and uracilâattached to each ribose-phosphate unit.
There are also numerous modified bases and sugars found in RNA that serve many different roles. Pseudouridine (Ψ), in which the linkage between uracil and ribose is changed from a CâN bond to a CâC bond, and ribothymidine (T), are found in various places (most notably in the TΨC loop of tRNA). Another notable modified base is hypoxanthine (a deaminated guanine base whose nucleoside is called inosine). Inosine plays a key role in the Wobble Hypothesis of the genetic code. There are nearly 100 other naturally occurring modified nucleosides, of which pseudouridine and nucleosides with 2'-O-methylribose are by far the most common. The specific roles of many of these modifications in RNA are not fully understood. However, it is notable that in ribosomal RNA, many of the post-translational modifications occur in highly functional regions, such as the peptidyl transferase center and the subunit interface, implying that they are important for normal function.
The most important structural feature of RNA that distinguishes it from DNA is the presence of a hydroxyl group at the 2'-position of the ribose sugar. The presence of this functional group enforces the C3'-endo sugar conformation (as opposed to the C2'-endo conformation of the deoxyribose sugar in DNA) that causes the helix to adopt the A-form geometry rather than the B-form most commonly observed in DNA. This results in a very deep and narrow major groove and a shallow and wide minor groove. A second consequence of the presence of the 2'-hydroxyl group is that in conformationally flexible regions of an RNA molecule (that is, not involved in formation of a double helix), it can chemically attack the adjacent phosphodiester bond to cleave the backbone.
Comparison with DNA
The most common nucleic acids are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). The main role of DNA is the long-term storage of genetic information. DNA is often compared to a blueprint, since it contains instructions for constructing other components of the cell, such as proteins and RNA molecules. The DNA segments that carry genetic information are called genes, but other DNA sequences have structural purposes or are involved in regulating the expression of genetic information. RNA, also, may serve more than one purpose, but it is most commonly identified as the intermediate between the DNA blueprint and the actual workings of the cell, serving as the template for the synthesis of proteins from the genetic information stored in DNA.
RNA and DNA differ in three main ways.
First, unlike DNA which is double-stranded, RNA is intrinsically a single-stranded molecule in most of its biological roles and has a much shorter chain of nucleotides. (While RNA is usually single-stranded, the RNA molecule also quite commonly forms double-helical regions where a given strand has folded back on itself. Double-stranded RNA is found also in certain viruses.)
Secondly, while DNA contains deoxyribose, RNA contains ribose. There is no hydroxyl group attached to the pentose ring in the 2' position in DNA, whereas RNA has two hydroxyl groups. These hydroxyl groups make RNA less stable than DNA because it is more prone to hydrolysis. (âDeoxyâ simply indicates that the sugar lacks an oxygen atom present in ribose, the parent compound.)
Thirdly, the complementary nucleotide to adenine is not thymine, as it is in DNA, but rather uracil, which is an unmethylated form of thymine.
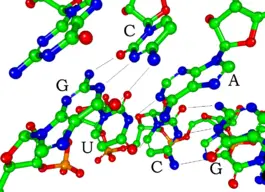
Most biologically active RNAs, including tRNA, rRNA, snRNAs, and other non-coding RNAs (such as the signal recognition particle(SRP) RNAs), contain extensively base paired regions that have folded together to form double stranded helices. Structural analysis of these RNAs reveals that they are highly structured with tremendous variety with collections of short helices packed together into structures much more akin to proteins than to DNA, which is usually limited to long double-stranded helices. Through such a variety of structures, RNAs can achieve chemical catalysis, like enzymes. For instance, determination of the structure of the ribosomeâan enzyme that catalyzes peptide bond formationârevealed that its active site is composed entirely of RNA.
Synthesis
Synthesis of RNA is usually catalyzed by an enzyme, RNA polymerase, using DNA as a template. Initiation of synthesis begins with the binding of the enzyme to a promoter sequence in the DNA (usually found "upstream" of a gene). The DNA double helix is unwound by the helicase activity of the enzyme. The enzyme then progresses along the template strand in the 3â -> 5â direction, synthesizing a complementary RNA molecule with elongation occurring in the 5â -> 3â direction. The DNA sequence also dictates where termination of RNA synthesis will occur (Nudler and Gottesman 2002).
There are also a number of RNA-dependent RNA polymerases as well that use RNA as their template for synthesis of a new strand of RNA. For instance, a number of RNA viruses (such as poliovirus) use this type of enzyme to replicate their genetic material (Hansen et al. 1997). Also, it is known that RNA-dependent RNA polymerases are required for the RNA interference pathway in many organisms (Ahlquist 2002).
Biological roles
RNA's great variety of possible structures and chemical properties permits it to perform a much greater diversity of roles than in the cell than DNA. Three principal types of RNA are involved in protein synthesis:
- Messenger RNA (mRNA) serves as the template for the synthesis of a protein. It carries information from DNA to the ribosome.
- Transfer RNA (tRNA) is a small chain of nucleotides that transfers a specific amino acid to a growing polypeptide chain at the ribosomal site of synthesis. It pairs the amino acid to the appropriate three-nucleotide codon on the mRNA molecule.
- Ribosomal RNA (rRNA) molecules are extremely abundant and make up at least 80 percent of the RNA molecules found in a typical eukaryotic cell. In the cytoplasm, usually three or four rRNA molecules combine with many proteins to perform a structural and essential catalytic role, as components of the ribosome.
RNA also may serve as a catalyst for reactions and as a genetic blueprint, rather than DNA, in various viruses. Some RNA, including tRNA and rRNA, is non-coding in that it is not translated into proteins.
Messenger RNA (mRNA)
Messenger RNA is RNA that carries information from DNA to the ribosome sites of protein synthesis in the cell. In eukaryotic cells, once mRNA has been transcribed from DNA, it is "processed" before being exported from the nucleus into the cytoplasm, where it is bound to ribosomes and translated into its corresponding protein form with the help of tRNA. In prokaryotic cells, which do not have nucleus and cytoplasm compartments, mRNA can bind to ribosomes while it is being transcribed from DNA. After a certain amount of time the message degrades into its component nucleotides, usually with the assistance of ribonucleases.
Non-coding RNA
RNA genes (also known as non-coding RNA or small RNA) are genes that encode RNA that is not translated into a protein. The most prominent examples of RNA genes are those coding for transfer RNA (tRNA) and ribosomal RNA (rRNA), both of which are involved in the process of translation. Two other groups of non-coding RNA are microRNAs (miRNA) which regulate the expression of genes through a process called RNA interference (RNAi), and small nuclear RNAs (snRNA), a diverse class that includes for example the RNAs that form spliceosomes that excise introns from pre-mRNA (Berg et al. 2002).
Transfer RNA (tRNA)
Transfer RNA is a small RNA chain of about 74-95 nucleotides that transfers a specific amino acid to a growing polypeptide chain at the ribosomal site of protein synthesis, during translation. It has sites for amino-acid attachment and an anticodon region for codon recognition that binds to a specific sequence on the messenger RNA chain through hydrogen bonding. It is a type of non-coding RNA.
Ribosomal RNA (rRNA)
Ribosomal RNA is the catalytic component of the ribosomes, the protein synthesis factories in the cell. Eukaryotic ribosomes contain four different rRNA molecules: 18S, 5.8S, 28S, and 5S rRNA. Three of the rRNA molecules are synthesized in the nucleolus, and one is synthesized elsewhere. rRNA molecules are extremely abundant and make up at least 80 percent of the RNA molecules found in a typical eukaryotic cell.
Catalytic RNA
Although RNA contains only four bases, in comparison to the twenty-odd amino acids commonly found in proteins, certain RNAs (called ribozymes) are still able to catalyze chemical reactions. These include cutting and ligating other RNA molecules, and also the catalysis of peptide bond formation in the ribosome.
Genetic blueprint in some viruses
Some viruses contain either single-stranded or double-stranded RNA as their source of genetic information. Retroviruses, for example, store their genetic information as RNA, though they replicate in their hosts via a DNA intermediate. Once in the host's cell, the RNA strands undergo reverse transcription to DNA in the cytosol and are integrated into the host's genome. Human immunodeficiency virus (or HIV) is a retrovirus thought to cause acquired immune deficiency syndrome (AIDS), a condition in which the human immune system begins to fail, leading to life-threatening opportunistic infections.
Double-stranded RNA (dsRNA) is RNA with two complementary strands, similar to the DNA found in all cells. dsRNA forms the genetic material of some viruses called dsRNA viruses. In eukaryotes, long RNA such as viral RNA can trigger RNA interference, where short dsRNA molecules called siRNAs (small interfering RNAs) can cause enzymes to break down specific mRNAs or silence the expression of genes. siRNA can also increase the transcription of a gene, a process called RNA activation (Doran 2007). siRNA is often confused with miRNA; siRNAs are double-stranded, whereas miRNAs are single-stranded.
RNA world hypothesis
The RNA world hypothesis proposes that the earliest forms of life relied on RNA both to carry genetic information (like DNA does now) and to catalyze biochemical reactions like an enzyme. According to this hypothesis, descendants of these early lifeforms gradually integrated DNA and proteins into their metabolism.
In the 1980s, scientists discovered that certain RNA molecules (called ribozymes) may function as enzymes, whereas previously only proteins were believed to have catalytic ability. Many natural ribozymes catalyze either their own cleavage or the cleavage of other RNAs, but they have also been found to catalyze the aminotransferase activity of the ribosome.
The discovery of ribozymes provides a possible explanation for how early RNA molecules might have first catalyzed their own replication and developed a range of enzymatic activities. Known as the RNA world hypothesis, this explanation posits that RNA evolved before either DNA or proteins from free-floating nucleotides in the early "primordial soup." In their function as enzymes, RNA molecules might have begun to catalyze the synthesis of proteins, which are more versatile than RNA, from amino acid molecules. Next, DNA might have been formed by reverse transcription of RNA, with DNA eventually replacing RNA as the storage form of genetic material. Although there are remaining difficulties with the RNA world hypothesis, it remains as a possible key to understanding the origin and development of the multi-functional nature of nucleic acids, the interconnectedness of life, and its common origins.
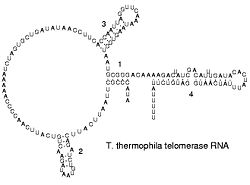
RNA secondary structures
The functional form of single stranded RNA molecules, just like proteins, frequently requires a specific tertiary structure. The scaffold for this structure is provided by secondary structural elements, which arise through the formation of hydrogen bonds within the interfolded molecule. This leads to several recognizable "domains" of secondary structure like hairpin loops, bulges, and internal loops. The secondary structure of RNA molecules can be predicted computationally by calculating the minimum free energies (MFE) structure for all different combinations of hydrogen bondings and domains (Mathews et al. 2004). There has been a significant amount of research directed at the RNA structure prediction problem.
History
Nucleic acids were discovered in 1868 by Johann Friedrich Miescher (1844-1895), who called the material 'nuclein' since it was found in the nucleus. It was later discovered that prokaryotic cells, which do not have a nucleus, also contain nucleic acids.
The role of RNA in protein synthesis had been suspected since 1939, based on experiments carried out by TorbjĂśrn Caspersson, Jean Brachet, and Jack Schultz. Hubert Chantrenne elucidated the messenger role played by RNA in the synthesis of proteins in ribosomes. Finally, Severo Ochoa discovered RNA, winning Ochoa the 1959 Nobel Prize for Medicine. The sequence of the 77 nucleotides of a yeast RNA was found by Robert W. Holley in 1964, winning Holley the 1968 Nobel Prize for Medicine. In 1976, Walter Fiers and his team at the University of Ghent determined the complete nucleotide sequence of bacteriophage MS2-RNA (Fiers et al. 1976).
List of RNA types
| Type | Function | Distribution |
|---|---|---|
| mRNA | Codes for protein | All cells |
| rRNA | Translation | All cells |
| tRNA | Translation | All cells |
| snRNA | RNA modification | All cells |
| snoRNA | RNA modification | All cells |
| miRNA | Gene regulation | Eukaryotes |
| piRNA | Gene regulation | Animal germline cells |
| siRNA | Gene regulation | Eukaryotes |
| Antisense mRNA | Preventing translation | Bacteria |
| tmRNA | Terminating translation | Bacteria |
| SRP RNA | mRNA tagging for export | All cells |
| Ribozyme | Catalysis | All cells |
| Transposon | Self-propagating | All cells |
| Viroid | Self-propagating | Infected plants |
In addition, the genome of many types of viruses consists of RNA, namely:
- Double-stranded RNA viruses
- Positive-sense RNA viruses
- Negative-sense RNA viruses
- Retroviruses
- Satellite viruses
ReferencesISBN links support NWE through referral fees
- Ahlquist, P. 2002. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science 296(5571): 1270-1273.
- Berg, J. M., J. L. Tymoczko, and L. Stryer. 2002. Biochemistry, 5th Edition. WH Freeman and Company. ISBN 0716746840.
- Doran, G. 2007. RNAi â Is one suffix sufficient? Journal of RNAi and Gene Silencing 3(1): 217-219.
- Fiers W et al. 1976. Complete nucleotide-sequence of bacteriophage MS2-RNA: Primary and secondary structure of replicase gene. Nature 260: 500-507.
- Hansen, J. L., A. M. Long, and S. C. Schultz. 1997. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5(8): 1109-1122. Retrieved December 7, 2007.
- Mathews, D. H., M. D. Disney, J. L. Childs, S. J. Schroeder, M. Zuker, and D. H. Turner. 2004. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. U. S. A. 101(19): 7287-7292. Retrieved December 6, 2007.
- Nudler, E., and M. E. Gottesman. 2002. Transcription termination and anti-termination in E. coli. Genes to Cells 7: 755-768. Retrieved December 7, 2007.
External links
All links retrieved December 7, 2022.
- RNAJunction Database: Extracted atomic models of RNA junction and kissing loop structures.
| Nucleic acids edit |
|---|
| Nucleobases: Adenine - Thymine - Uracil - Guanine - Cytosine - Purine - Pyrimidine |
| Nucleosides: Adenosine - Uridine - Guanosine - Cytidine - Deoxyadenosine - Thymidine - Deoxyguanosine - Deoxycytidine |
| Nucleotides: AMP - UMP - GMP - CMP - ADP - UDP - GDP - CDP - ATP - UTP - GTP - CTP - cAMP - cGMP |
| Deoxynucleotides: dAMP - dTMP - dUMP - dGMP - dCMP - dADP - dTDP - dUDP - dGDP - dCDP - dATP - dTTP - dUTP - dGTP - dCTP |
| Nucleic acids: DNA - RNA - LNA - PNA - mRNA - ncRNA - miRNA - rRNA - siRNA - tRNA - mtDNA - Oligonucleotide |
Credits
New World Encyclopedia writers and editors rewrote and completed the Wikipedia article in accordance with New World Encyclopedia standards. This article abides by terms of the Creative Commons CC-by-sa 3.0 License (CC-by-sa), which may be used and disseminated with proper attribution. Credit is due under the terms of this license that can reference both the New World Encyclopedia contributors and the selfless volunteer contributors of the Wikimedia Foundation. To cite this article click here for a list of acceptable citing formats.The history of earlier contributions by wikipedians is accessible to researchers here:
The history of this article since it was imported to New World Encyclopedia:
Note: Some restrictions may apply to use of individual images which are separately licensed.