An enzyme is a biological catalyst that regulates the rate of a chemical reaction in a living organism. Most enzymes are proteins, though certain nucleic acids, called ribozymes, are also capable of catalytic activity.
Enzymes are essential to sustain life because most chemical reactions in biological cells, such as the digestion of food, would occur too slowly or would lead to different products without the activity of enzymes. Most inherited human diseases result from a genetic mutation, overproduction, or deficiency of a single critical enzyme. For example, lactose intolerance, the inability to digest significant amounts of lactose, which is the major sugar found in milk, is caused by a shortage of the enzyme lactase.
For an enzyme to be functional, it must fold into a precise three-dimensional shape. How such a complex folding can take place remains a mystery. A small chain of 150 amino acids making up an enzyme has an extraordinary number of possible folding configurations: if it tested 1012 different configurations every second, it would take about 1026 years to find the right one (Lewis 2005). Yet, a denatured enzyme can refold within fractions of a second and then precisely react in a chemical reaction. To some, it suggests that quantum effects are at work even at the large distances (by atomic standards) spanned by a protein molecule. At least, it demonstrates a stunning complexity and harmony in the universe.
While all enzymes have a biological role, some enzymes are also used commercially. For instance, many household cleaners use enzymes to speed up the breakdown of protein or starch stains on clothes.
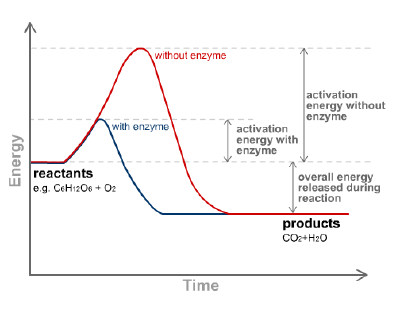
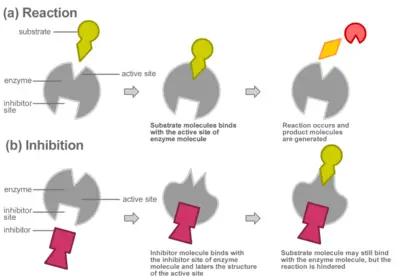
Like all catalysts, enzymes work to lower the activation energy of a reaction, or the initial energy input necessary for most chemical reactions to occur. Heat cannot be added to a living system, so enzymes provide an alternate pathway: they bond with a substrate (the substance involved in the chemical reaction) to form a ‚Äútransition state,‚ÄĚ an unstable intermediate complex that requires less energy for the reaction to proceed. Like any catalyst, the enzyme remains unaltered by the completed reaction and can therefore continue to interact with substrates. Enzymes may speed up reactions by a factor of many millions.
Enzymes can be affected by molecules that increase their activity (activators) or decrease their activity (inhibitors). Many drugs act by inhibiting enzymes. Aspirin works by inhibiting COX-1 and COX-2, the enzymes that produce prostaglandin, a hormonal messenger that signals inflammation. By inhibiting the activity of these enzymes, aspirin suppresses our experience of pain and inflammation.
The structure of enzymes
Enzyme structure is important because it determines the enzyme's particular function in the body. Enzymes (and other proteins) are composed of amino acid chains called polypeptide chains. The linear sequence of amino acids determines the characteristic folding of the chains into a three-dimensional structure. An enzyme might contain only one polypeptide chain, typically linking one hundred or more amino acids, or it might consist of several polypeptide chains that act together as a unit.
Most enzymes are larger than the substrates on which they act. Only a very small portion of the enzyme, approximately ten amino acids, comes into direct contact with the substrate(s). This region, where the binding of the substrate(s) and the reaction occur, is known as the active site of the enzyme.
Specificity
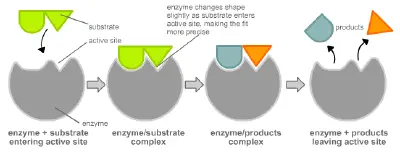
Enzymes are usually specific, or unique, to the reactions they catalyze and the substrates that are involved in these reactions. An enzyme combines with its substrate(s) to form a short-lived enzyme-substrate complex. There are two models to explain how the binding of enzyme and substrate occurs: the "lock and key" model and induced fit.
"Lock and key" model
To account for the specificity of enzymes, Emil Fischer proposed that the enzyme had a particular shape into which the substrate(s) fit exactly. This model of exact fit, introduced in the 1890s, is often referred to as the "lock and key" model, because the enzyme binding to a substrate is analogous to the specific fit of a lock into a key.
Induced fit model
In 1958, Daniel Koshland suggested a modification to the "lock and key" model. Unlike keys, enzymes are rather flexible structures. The active site of an enzyme can be modified as the substrate interacts with the enzyme, creating an "induced fit" between enzyme and substrate. The amino acids side chains that make up the active site are molded into a precise shape, which enables the enzyme to perform its catalytic function. In some cases, the substrate molecule changes shape slightly as it enters the active site.
Enzyme cofactors
Some enzymes do not need any additional components to exhibit full activity. However, others require non-protein molecules to be bound to the complex for efficient activity. Cofactors can be either inorganic (e.g., metal ions and iron-sulfur clusters) or organic compounds, which are also known as coenzymes.
Most cofactors are not covalently bound to an enzyme, but are closely associated. However, some cofactors known as prosthetic groups are tightly bound to the enzyme through covalent bonds.
Most cofactors are either regenerated or chemically unchanged at the end of the reactions. Many cofactors are vitamin-derivatives. They serve as carriers during the reaction to transfer electrons, atoms, or functional groups from an enzyme to a substrate. Common examples include NAD and NADP, which are involved in electron transfer, and coenzyme A, which is involved in the transfer of acetyl groups.
How enzymes catalyze reactions
A reaction catalyzed by enzymes must be spontaneous; that is, having a natural tendency to occur without needing an external push. (Thermodynamically speaking, the reaction must contain a net negative Gibbs free energy.) In other words, the reaction would run in the same direction without the enzyme, but would occur at a significantly slower rate. For example, the breakdown of food particles such as carbohydrates into smaller sugar components occurs spontaneously, but the addition of enzymes such as amylases in our saliva makes the reaction occur quickly.
Enzymes can pair two or more reactions, so that a spontaneous reaction can be used to drive an unfavorable one. For example, the cleavage of the high-energy compound ATP is often used to power other, energetically unfavorable chemical reactions, such as the building of proteins.
Regulation of enzyme activity
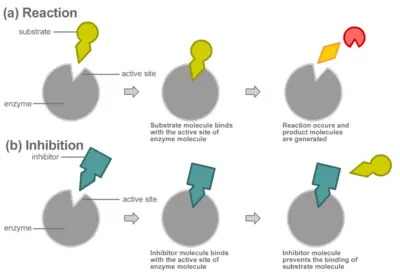
Compounds called inhibitors can decrease enzyme reaction rates through competitive or non-competitive inhibition.
In competitive inhibition, the inhibitor binds directly to the active site as shown, preventing the binding of substrate. The substrate and inhibitor thus "compete" for the active site of the enzyme.
Non-competitive inhibitors do not bind to the active site; rather, they bind to other parts of the enzyme, which can be remote from the active site. The extent of inhibition depends entirely on the inhibitor concentration and will not be affected by the substrate concentration. For example, the poison cyanide combines with the copper prosthetic groups of the enzyme cytochrome c oxidase to inhibit cellular respiration. This type of inhibition is typically irreversible, meaning that the enzyme will no longer function after interacting with the inhibitor.
Some non-competitive inhibitors work by physically blocking the active site. Others bind to the enzyme in a way that alters the three-dimensional structure of the enzyme (its conformation); the change in the enzyme's structure distorts the active site, disabling the enzyme from binding with substrate. In this second form of noncompetitive inhibition, called allosteric inhibition, the inhibitor binds to an allosteric site, changing the shape of the enzyme molecule in a way that prevents it from reacting with the substrate.
Allosteric control
Allosteric inhibitors are often used to regulate metabolic pathways, in which several enzymes work together in a specific order. In a metabolic pathway, one enzyme takes the product of another enzyme as a substrate. After the catalytic reaction, the product is then passed on to another enzyme. The end product(s) of such a pathway are often allosteric inhibitors for one of the first enzymes of the pathway (usually the first irreversible step, called the committed step), thus regulating the amount of end product made by the pathways. This regulatory process is called negative feedback, because the amount of the end product produced is regulated by its own concentration.
Allosteric molecules can also activate or increase the activity of enzymes by changing the shape of the enzyme's active site in order to facilitate interaction with a substrate. This allosteric control of enzymatic action helps to maintain a stable internal environment in living organisms, by stimulating the production of supplies when needed and preventing the excess manufacture of end products once the demand has been met.
Enzyme-naming conventions
Enzymes are known for their specificity; that is, they often interact with only one substrate to catalyze a particular reaction. Thus, enzymes have often been named by adding the suffix -ase to the name of the substrate (e.g., lactase is the enzyme that catalyzes the breakdown of lactose). Not all enzymes have been named in this manner, so a more formal method of nomenclature has been developed to classify enzymes.
The International Union of Biochemistry and Molecular Biology has developed a nomenclature for enzymes, called EC numbers. The EC number describes each enzyme using a sequence of four numbers, preceded by "EC." The first number broadly classifies the enzyme based on how it functions to catalyze a reaction.
Under this system, enzymes are broadly organized into six major categories, based on the types of reactions they catalyze:
- EC 1 Oxidoreductases catalyze oxidation/reduction reactions, which involve electron transfer.
- EC 2 Transferases transfer a chemical group called a functional group (e.g., a methyl or phosphate group) from one substance to another.
- EC 3 Hydrolases catalyze the cleavage of chemical bonds through the addition of a water molecule hydrolysis.
- EC 4 Lyases cleave various bonds by means other than hydrolysis and oxidation.
- EC 5 Isomerases transfer a group within a single molecule to form an isomer.
- EC 6 Ligases join two molecules with covalent bonds.
Related Topics
Etymology and history
The word enzyme derives from the Greek ő≠őĹő∂ŌÖőľőŅ, √©nsymo, which comes from √©n ("at" or "in") and simo ("leaven" or "yeast"). Although the leavening of bread and fermentation of wine had been practiced for centuries, these processes were not understood to be the result of enzyme activity until the late nineteenth century.
Studying the fermentation of sugar to alcohol by yeast, Louis Pasteur came to the conclusion that this fermentation was catalyzed by ferments in the yeast, which were thought to function only in the presence of living organisms. However, in 1897, Hans and Eduard Buchner inadvertently used yeast extracts to ferment sugar, despite the absence of living yeast cells. They were interested in making extracts of yeast cells for medical purposes, and, as one possible way of preserving them, they added large amounts of sucrose to the extract. To their surprise, they found that the sugar was fermented, even though there were no living yeast cells in the mixture. The term "enzyme" was used to describe the substance(s) in yeast extract that brought about the fermentation of sucrose. It was not until 1926 that the first enzyme was obtained in pure form.
Enzyme kinetics
In 1913 Leonor Michaelis and Maud Menten proposed a quantitative theory of enzyme kinetics, which is referred to as Michaelis-Menten kinetics. Their work was further developed by G. E. Briggs and J. B. S. Haldane, who derived numerous kinetic equations that are still widely used today.
Enzymes can perform up to several million catalytic reactions per second. To determine the maximum speed of an enzymatic reaction, the substrate concentration is increased until a constant rate of product formation is achieved. This rate is the maximum velocity (Vmax) of the enzyme. In this state, all enzyme active sites are saturated with substrate; that is, they are all engaged in converting substrate to product.
However, Vmax is only one kinetic parameter that interests biochemists. They also want to be able to calculate the amount of substrate needed to achieve a given rate of reaction. This amount can be expressed by the Michaelis-Menten constant (Km), which is the substrate concentration required for an enzyme to reach one half its maximum velocity. Each enzyme has a characteristic Km for a given substrate.
The efficiency of an enzyme can be expressed in terms of kcat/Km. The quantity kcat, also called the turnover number, incorporates the rate constants for all steps in the reaction, and is the quotient of Vmax and the total enzyme concentration. kcat/Km is a useful quantity for comparing the relative efficiencies of different enzymes, or the same enzyme interacting with different substrates, because it takes both affinity and catalytic ability into consideration. The theoretical maximum for kcat/Km, called the diffusion limit, is about 108 to 109 (M-1 s-1). At this point, every collision of the enzyme with its substrate will result in catalysis, and the rate of product formation is not limited by the reaction rate but by the diffusion rate. Enzymes that reach this kcat/Km value are called catalytically perfect or kinetically perfect. Example of such enzymes include triose-phosphate isomerase (or TIM), carbonic anhydrase, acetylcholinesterase, catalase, fumarase, ß-lactamase, and superoxide dismutase.
Industrial applications
Below are some common applications of enzymes, which have played an increased role in industrial processes since the scientific understanding of their catalytic function in the late nineteenth century:
Proteases, which function in the breakdown of the bonds between the amino acids that constitute protein molecules, are used in biological detergents to help with the removal of protein stains. Rennin, a type of protease that is derived from the stomachs of young ruminant animals (calves, lambs), is used to split protein during the manufacture of cheese. Another type of protease called trypsin is used to pre-digest baby foods.
- Amylase, a digestive enzyme used in the breakdown of carbohydrates, helps to remove resistant starch residues in dishwashing detergents. Fungal-alpha amylase enzymes catalyze the breakdown of starch in flour into its component sugars; they are used in the production of white bread, buns, and rolls.
- The brewing industry utilizes a variety of enzymes released from the malt (often the grain barley) during the mashing stage of beer production, in which the barley and water are combined and heated. These enzymes, which include amylases, glucanases, and proteases, degrade starches and proteins in the malt to produce simple sugar, amino acids and peptides that enhance fermentation.
ReferencesISBN links support NWE through referral fees
- Briggs, G. E. & J. B. S. Haldane. 1925. A note on the kinetics of enzyme action, Biochem. J. 19:339-339.
- Cha, Y., C. J. Murray, & J. P. Klinman. 1989. Science 243: 1325-1330.
- Koshland, D. 1959. The Enzymes. New York: Academic Press.
- Lewis, R. L. 2005. Do Proteins Teleport in an RNA World. New York: International Conference on the Unity of the Sciences.
- Michaelis, L. and M. Menten. 1913. Die Kinetik der Invertinwirkung, Biochem. Z. 49:333-369.
- Perutz, M. 1967. Proc. Roy. Soc. 167: 448.
- Volkenshtein, M.V., R.R. Dogonadze, A.K. Madumarov, Z.D. Urushadze, & Yu.I. Kharkats. 1972. Theory of Enzyme Catalysis, Molekuliarnaya Biologia. 431-439 (In Russian, English summary).
External links
All links retrieved February 13, 2024.
- Enzyme Nomenclature and Kinetics International Union of Biochemistry and Molecular Biology
- Enzymes Cleveland Clinic
- Enzymes: How they work and what they do Medical News Today
Credits
New World Encyclopedia writers and editors rewrote and completed the Wikipedia article in accordance with New World Encyclopedia standards. This article abides by terms of the Creative Commons CC-by-sa 3.0 License (CC-by-sa), which may be used and disseminated with proper attribution. Credit is due under the terms of this license that can reference both the New World Encyclopedia contributors and the selfless volunteer contributors of the Wikimedia Foundation. To cite this article click here for a list of acceptable citing formats.The history of earlier contributions by wikipedians is accessible to researchers here:
The history of this article since it was imported to New World Encyclopedia:
Note: Some restrictions may apply to use of individual images which are separately licensed.