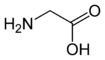
Glycine
| |
Glycine | |
| Systematic (IUPAC) name | |
| aminoethanoic acid | |
| Identifiers | |
| CAS number | 56-40-6 |
| PubChem | 750 |
| Chemical data | |
| Formula | C2H5NO2 |
| Mol. weight | 75.07 |
| SMILES | NCC(O)=O |
| Complete data | |
Glycine is the simplest of the α-amino acids found in proteins in terms of molecular structure. It is one of the 20 most common, natural, "proteinogenic" (literally, protein building) amino acids (standard amino acids. Glycine is unique among these in that is not optically active; that is, it does not have occur in two possible optical isomers, called D and L.
While glycine is necessary for normal functioning in humans, it is categorized as a non-essential amino acid since it does not have to be taken in with the diet, but can be synthesized by the human body from other compounds through chemical reactions.
Most proteins contain only small quantities of glycine. A notable exception is collagen, which contains about one-third glycine. Glycine is a major component of silk, along with alanine, as well as gelatin.
Glycine's three letter code is GLY, its one letter code is G, and its codons are GGU, GGC, GGA and GGG (IUPAC-IUB 1983). Its systematic name is aminoethanoic acid. systematic name
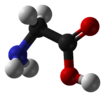
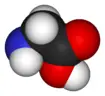
Structure
Note:In biochemistry, the term amino acid is frequently used to refer specifically to alpha amino acids: those amino acids in which the amino and carboxylate groups are attached to the same carbon, the so-called α–carbon (alpha carbon). The general structure of these alpha amino acids is:
R
|
H2N-C-COOH
|
H
where R represents a side chain specific to each amino acid.
Most amino acids occur in two possible optical isomers, called D and L. The L amino acids represent the vast majority of amino acids found in proteins. Glycine, however, because of its simple structure and two hydrogen atoms at the α carbon, does not have D- and L-stereoisomers. It is unqiue among the 20 standard amino acids in not being optically active.
Glycine is the smallest α-amino acid, is structurally simple (having a side chain of just hydrogen), rotates easily, and adds flexibility to the protein chain. It is able to fit into the tightest spaces, e.g., the triple helix of collagen. Because of its structural simplicity, this compact amino acid tends to be evolutionarily conserved in, for example, cytochrome c, myoglobin, and hemoglobin.
Glycine has the chemical formula CH2(NH2)-COOH, or more generally, C2H5NO2.
Biosynthesis
Glycine is not essential to the human diet, since it is synthesized in the body. It is biosynthesized from the amino acid serine. The enzyme serine hydroxymethyl transferase catalyses this transformation:[1]
- HO2CCH(NH2)CH2OH + H2folate → HO2CCH2NH2 + CH2-folate + H2O
Physiological function
As a biosynthetic intermediate
Glycine is a building block to numerous species. Aminolevulinic acid, the key precursor to porphyrins is biosynthesized from glycine and succinoyl coenzyme A. Glycine provides the central C2N subunit of all purines.[1]
As a neurotransmitter
Glycine is an inhibitory neurotransmitter in the central nervous system, especially in the spinal cord, brainstem, and retina. When glycine receptors are activated, chloride enters the neuron via ionotropic receptors, causing an Inhibitory postsynaptic potential (IPSP). Strychnine is an antagonist at ionotropic glycine receptors. Glycine is a required co-agonist along with glutamate for NMDA receptors. In contrast to the inhibitory role of glycine in the spinal cord, this behaviour is facilitated at the (NMDA) glutaminergic receptors which are excitatory. The LD50 of glycine is 7930 mg/kg in rats (oral),Template:Ref N and it usually causes death by hyperexcitability.
Presence in the interstellar medium
In 1994 a team of astronomers at the University of Illinois, led by Lewis Snyder, claimed that they had found the glycine molecule in space. It turned out that, with further analysis, this claim could not be confirmed. Nine years later, in 2003, Yi-Jehng Kuan from National Taiwan Normal University and Steve Charnley claimed that they detected interstellar glycine toward three sources in the interstellar medium Template:Ref N. They claimed to have identified 27 spectral lines of glycine utilizing a radio telescope. According to computer simulations and lab-based experiments, glycine was probably formed when ices containing simple organic molecules were exposed to ultraviolet light Template:Ref N.
In October 2004, Snyder and collaborators reinvestigated the glycine claim in Kuan et al. (2003). In a rigorous attempt to confirm the detection, Snyder showed that glycine was not detected in any of the three claimed sources Template:Ref N.
Should the glycine claim be substantiated, the finding would not prove that life exists outside the Earth, but certainly makes that possibility more plausible by showing that amino acids can be formed in the interstellar medium. The finding would also indirectly support the idea of panspermia, the theory that life was brought to Earth from space. As the simplest of amino acids, it seems one of the most like to be detected in the interstellar medium.
Silk
Silk
Alanine is a key component in spider silk. Spider silk is a remarkably strong material, with a tensile strength is comparable to that of high-grade steel (Shao and Vollrath 2002). Spider silk is so strong that it has been said that a circular web, similar in all ways to that found in nature but the size of a football field, could stop a commercial jetliner in mid flight (Henke 2007), and yet it is so light that a single strand strand long enough to circle the earth would weigh less than 16 ounces (460 g). The particular arrangement of the amino acids reveals the complex coordination in nature, a harmony that has existed for millions of years and which scientists now are studying in hope of learning how to create such a strong and yet elastic fiber.
Spider dragline silk is made up of the protein fibroin, which is a combination of the proteins spidroin 1 and spidroin 2. The bulk of these proteins are made up of alanine (Ala) and glycine (Gly), with the remaining components mostly the amino acids proline (Pro), tyrosine (Tyr), arginine (Arg), glutamine (Gln), serine (Ser), and leucine (Leu) (UB 2007). Spidroin 1 and 2 are made up of polyalanine regions with about 4 to 9 alanine monomers in a block (van Beek et al. report approximately 8 monomers) and glycine rich areas with a sequence of five amino acids continuously repeated, such as Gly-Pro-Gly-Gln-Gln (van Beek et al. 2002; UB 2007).
The general trend in spider silk structure thus is a sequence of amino acids (usually alternating glycine and alanine, or alanine alone) that self-assemble into a beta sheet conformation. These "Ala rich" blocks are separated by segments of amino acids with bulky side-groups. The beta sheets stack to form crystals, whereas the other segments form amorphous domains. It is the interplay between the hard crystalline segments, and the elastic semi- amorphous regions, that gives spider silk its extraordinary properties. The fact that the major amino acids in spider silk are the two smallest amino acids, and lack bulky side groups, allows them to pack together tightly (UB 2007).
The glycine-rich regions give spider silk its elasticity, as each sequence of five amino acids is followed by a 180 degree turn, resulting in a spiral. Capture silk is the most elastic, with about 43 repeats on average, and can extend 2 to 4 times its original length, while dragline silk only repeats about 9 times and can extend about 30% of original length (UB 2007).
ReferencesISBN links support NWE through referral fees
- Template:Note N Kuan YJ, Charnley SB, Huang HC, et al. (2003) Interstellar glycine. ASTROPHYS J 593 (2): 848-867
- Template:Note N Snyder LE, Lovas FJ, Hollis JM, et al. (2005) A rigorous attempt to verify interstellar glycine. ASTROPHYS J 619 (2): 914-930
- Template:Note NSafety (MSDS) data for glycine. The Physical and Theoretical Chemistry Laboratory Oxford University (2005). Retrieved 2006-11-01.
- Dawson, R.M.C., Elliott, D.C., Elliott, W.H., and Jones, K.M., Data for Biochemical Research (3rd edition), pp. 1-31 (1986)
- Template:Note N www.newscientist.com
Notes: * Doolittle, R. F. 1989. Redundancies in protein sequences. In G. D. Fasman, ed., Prediction of Protein Structures and the Principles of Protein Conformation. New York: Plenum Press. ISBN 0306431319.
- Henke, B. 2007. The webs they weave. Poststar.com Sunday, June 10, 2007. Retrieved June 15, 2007.
- International Union of Pure and Applied Chemistry and International Union of Biochemistry and Molecular Biology (IUPAC-IUB) Joint Commission on Biochemical Nomenclature. 1983. Nomenclature and symbolism for amino acids and peptides: Recommendations on organic & biochemical nomenclature, symbols & terminology. IUPAC-IUB. Retrieved June 14, 2007.
- Kendall, E. C., and B. F. McKenzie. 1941. dl-Alanine. Organic Syntheses 1: 21.
- Shao, Z., and F. Vollrath. 2002. Surprising strength of silkworm silk. Nature 418: 741.
- University of Bristol, School of Chemistry (UB). 2007. Spider silk: Chemical structure. University of Bristol. Retrieved June 14, 2007.
- van Beek, J. D., S. Hess, F. Vollrath, and B. H. Meier. 2002. The molecular structure of spider dragline silk: Folding and orientation of the protein backbone. Proc Natl Acad Sci USA 99(16): 10266-10271. Retrieved June 14, 2007.
External links
Template:ChemicalSources
| Major families of biochemicals | ||
| Peptides | Amino acids | Nucleic acids | Carbohydrates | Nucleotide sugars | Lipids | Terpenes | Carotenoids | Tetrapyrroles | Enzyme cofactors | Steroids | Flavonoids | Alkaloids | Polyketides | Glycosides | ||
| Analogues of nucleic acids: | The 20 Common Amino Acids | Analogues of nucleic acids: |
| Alanine (dp) | Arginine (dp) | Asparagine (dp) | Aspartic acid (dp) | Cysteine (dp) | Glutamic acid (dp) | Glutamine (dp) | Glycine (dp) | Histidine (dp) | Isoleucine (dp) | Leucine (dp) | Lysine (dp) | Methionine (dp) | Phenylalanine (dp) | Proline (dp) | Serine (dp) | Threonine (dp) | Tryptophan (dp) | Tyrosine (dp) | Valine (dp) | ||
Credits
New World Encyclopedia writers and editors rewrote and completed the Wikipedia article in accordance with New World Encyclopedia standards. This article abides by terms of the Creative Commons CC-by-sa 3.0 License (CC-by-sa), which may be used and disseminated with proper attribution. Credit is due under the terms of this license that can reference both the New World Encyclopedia contributors and the selfless volunteer contributors of the Wikimedia Foundation. To cite this article click here for a list of acceptable citing formats.The history of earlier contributions by wikipedians is accessible to researchers here:
The history of this article since it was imported to New World Encyclopedia:
Note: Some restrictions may apply to use of individual images which are separately licensed.
- ↑ 1.0 1.1 Nelson, D. L.; Cox, M. M. "Lehninger, Principles of Biochemistry" 3rd Ed. Worth Publishing: New York, 2000. ISBN 1-57259-153-6.