Lysosome
Lysosome is an organelle of eukaryotic cells that contains hydrolytic enzymes active under acidic conditions and involved in intracellular digestion. This membrane-bound sub-cellular structure contains a unique collection of about 40 hydrolytic enzymes, including proteases, nucleases, lipases, glycosidases, phospholipases, phosphatases, and sulfates, all of which are acid hydrolases, optimally active near pH 5 (Alberts et al. 1989). They digest excess or worn-out organelles, food particles, and engulfed viruses or bacteria.
Lysosomes are part of the intricate coordination of diverse cellular components, each type of organelle and sub-cellular component fulfilling different but vital functions for the cell, all working together in harmony. Lysosomes utilize unique enzymes and various pathways (phagocytosis, endocytosis, autophagy) in fulfilling their particular role, designed to degrade only those macromolecules targeted for destruction. The unique surrounding membrane of the lysosome keeps these enzymes out of the cystosol, where they might damage proteins and nucleic acids, while the acid dependence of the enzymes provides some protection even in leakage given the higher pH (around 7.2) of the cytosol (Alberts et al. 1989).
The name lysosome derives from the Greek words lysis, which means dissolution or destruction, and soma, which means body. They are frequently nicknamed "suicide-bags" or "suicide-sacs" by cell biologists due to their role in autolysis (destruction of a cell through the action of its own enzymes). Lysosomes were discovered by the Belgian cytologist Christian de Duve in 1949.
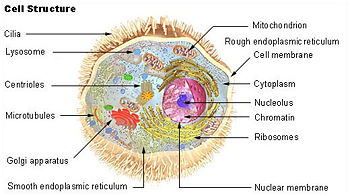
Overview
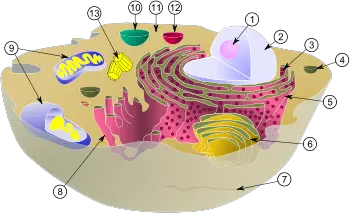
(1) nucleolus
(2) nucleus
(3) ribosomes (little dots)
(4) vesicle
(5) rough endoplasmic reticulum (ER)
(6) Golgi apparatus
(7) Cytoskeleton
(8) smooth ER
(9) mitochondria
(10) vacuole
(11) cytoplasm
(12) lysosome
(13) centrioles within centrosome
Lysosomes are membranous bags of hydrolytic enzymes and the principle site of intracellular digestion, being used for controlled digestion of macromolecules (Alberts et al. 1989). Lysomes are created by the addition of hydrolytic enzymes to early endosomes from the Golgi apparatus.
Lysosomes are very diverse in size and shape. This heterogeneity contrasts with the relatively uniform morphology of most cellular organelles, and reflects the wide variety of digestive functions of acid hydrolases, including digestion of phagocytosed microorganisms and intra- and extracellular debris, as well as principal site of cholesterol assimilation from endocytosed serum lipoprotein (Alberts et al. 1989). Lysosomes sometimes are considered a heterogeneous collection of distinct organelles united by having a high amount of hydrolytic enzymes (Alberts et al. 1989).
Lysosomes have a unique surrounding membrane. It contains transport proteins such that the final products of the digestion can pass out of the lysosome and be utilized by the cell or excreted (Alberts et al. 1989). The lysosomal membrane also contains a H+ pump to maintain an acidic pH (Alberts et al. 1989). Thus, the membrane surrounding a lysosome allows the digestive enzymes to work at the 4.5 pH they require. The proteins in the lysosomal membrane generally are unusually highly glycosylated, perhaps helping to protect them from the proteases inside the lysosome (Alberts et al. 1989).
Lysosomes fuse with vacuoles and dispense their enzymes into the vacuoles, digesting their contents.
At pH 4.8, the interior of the lysosomes is more acidic than the cytosol (pH 7.2). The lysosome's single membrane stabilizes the low pH by pumping in protons (H+) from the cytosol via proton pumps and chloride ion channels. The membrane also protects the cytosol, and therefore the rest of the cell, from the degradative enzymes within the lysosome. For this reason, should a lysosome's acid hydrolases leak into the cytosol, their potential to damage the cell will be reduced, because they will not be at their optimum pH.
Enzymes
Some important enzymes in lysosomes include:
- Lipase, which digests lipids
- Carbohydrases, which digest carbohydrates (e.g., sugars)
- Proteases, which digest proteins
- Nucleases, which digest nucleic acids
- phosphoric acid monoesters.
Glycosidases are enzymes involved in the degradation of carbohydrate structures, catalyzing the hydrolysis of the glycosidic linkage to generate two smaller sugars. A phospholipase is an enzyme that converts phospholipids into fatty acids and other lipophilic substances. A phosphatase is an enzyme that removes a phosphate group from its substrate by hydrolyzing phosphoric acid monoesters into a phosphate ion and a molecule with a free hydroxyl group. Sulfatases are esterase enzymes, which remove sulfate from a variety of substrates by hydrolyzing various sulfate esters.
Lysosomal enzymes are synthesized in the cytosol and the endoplasmic reticulum, where they receive a mannose-6-phosphate tag that targets them for transport to the lysosome. Aberrant lysosomal targeting causes inclusion-cell disease, whereby enzymes do not properly reach the lysosome, resulting in accumulation of waste within these organelles.
Functions
Lysosomes function in the digestion of macromolecules from three principle pathways: phagocytosis (ingestion of other dying cells or larger extracellular material); endocytosis (where receptor proteins are recycled from the cell surface); and autophagy (wherein old or unneeded organelles or proteins, or microbes that have invaded the cytoplasm are delivered to the lysosome).
In phagocytosis, large particles and microorganisms are engulfed by cells such as macrophages and neutrophis to form a phagosome, which fuses with a lysososme to result in the digestion (Alberts et al. 1989).
In endocystosis, ingested macromolecules sequestered in vesicles (having been progressively enclosed by a small portion of the plasma membrane), are delivered to peripheral endosomes and move to perinuclear endosomes, endolysososmes, and finally to lysosomes, where they are degraded (Alberts et al. 1989).
As an example of autophagy, the average mitochondrion in a liver cell exists about ten days. Electron microscopes of normal cells reveal lysosomes containing, and presumably digesting, mitochondrions. This process appears to involve enclosure of the organelle by membranes derived from the endoplasmic reticulum, and this resultant autophagosome then fuses with a lysosome to result in digestion of the contents (Alberts et al. 1989).
Autophagy may also lead to autophagic cell death, a form of programmed self-destruction, or autolysis, of the cell, which means that the cell is digesting itself through the action of its own enzymes. Autolysis in initiated by the lysosomes releasing the digestive enzymes they contain out into the cytoplasm; the cell then, in effect, starts to digests itself.
Other functions of lysosomes include digesting foreign bacteria (or other forms of waste) that invade a cell and helping repair damage to the plasma membrane by serving as a membrane patch, sealing the wound. In the past, lysosomes were thought to kill cells that were no longer wanted, such as those in the tails of tadpoles or in the web from the fingers of a three- to six-month-old fetus. While lysosomes digest some materials in this process, it is actually accomplished through programmed cell death, called apoptosis (Kimball 2007; Mader and Pendarvis 2007).
Clinical relevance
There are a number of illnesses that are caused by the malfunction of the lysosomes or one of their digestive proteins, such as Tay-Sachs disease and Pompe disease. These are caused by a defective or missing digestive protein, which leads to the accumulation of substrates within the cell, impairing metabolism.
Tay-Sachs disease is a condition is caused by insufficient activity of an enzyme called hexosaminidase A that catalyzes the biodegradation of fatty acid derivatives known as gangliosides. Hexasaminidase A is a vital hydrolytic enzyme, found in the lysosomes, that breaks down lipids. When Hexasaminidase A is no longer functioning properly, the lipids accumulate in the brain and cause problems.
Pompe disease, also known as glycogen storage disease type II, is a rare, autosomal recessive metabolic disorder caused by a deficiency in the enzyme acid maltase, which is needed to break down glycogen, a stored form of sugar used for energy. It is the only glycogen storage disease with a defect in lysosomal metabolism.
In the broad sense, illness caused by malfunctions related to the lyssosome or their digestive proteins can be classified as mucopolysaccharidoses, GM2 gangliosidoses, lipid storage disorders, glycoproteinoses, mucolipidoses, or leukodystrophies.
Additional images
ReferencesISBN links support NWE through referral fees
This article contains material from the Science Primer published by the NCBI, which, as a US government publication, is in the public domain at http://www.ncbi.nlm.nih.gov/About/disclaimer.html.
- Alberts, B., D. Bray, J. Lewis, M. Raff, K. Roberts, and J. D. Watson. Molecular Biology of the Cell, 2nd edition. New York: Garland Publishing, 1989. ISBN 0824036956.
- Bainton, D. 1981. The discovery of lysosomes. J. Cell Biology 91: 66s-76s.
- Kimball, J. 2007. Lysosomes and peroxisomes Kimball's Biology Pages.
- Mader, S. S., and M. P. Pendarvis. 2007. Biology. Boston: McGraw-Hill Higher Education. ISBN 9780072464634.
| Organelles of the cell |
|---|
| Acrosome | Chloroplast | Cilium/Flagellum | Centriole | Endoplasmic reticulum | Golgi apparatus | Lysosome | Melanosome | Mitochondrion | Myofibril | Nucleus | Parenthesome | Peroxisome | Plastid | Ribosome | Vacuole | Vesicle |
Credits
New World Encyclopedia writers and editors rewrote and completed the Wikipedia article in accordance with New World Encyclopedia standards. This article abides by terms of the Creative Commons CC-by-sa 3.0 License (CC-by-sa), which may be used and disseminated with proper attribution. Credit is due under the terms of this license that can reference both the New World Encyclopedia contributors and the selfless volunteer contributors of the Wikimedia Foundation. To cite this article click here for a list of acceptable citing formats.The history of earlier contributions by wikipedians is accessible to researchers here:
- Lysosome  history
- Autolysis_(biology)  history
- Glycoside_hydrolase  history
- Phospholipase  history
- Phosphatase  history
- Sulfatase  history
- Glycogen_storage_disease_type_II  history
- Tay-Sachs_disease  history
The history of this article since it was imported to New World Encyclopedia:
Note: Some restrictions may apply to use of individual images which are separately licensed.