Difference between revisions of "Plasmid" - New World Encyclopedia
Rosie Tanabe (talk | contribs) m |
|||
| (17 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
| − | {{ | + | {{Approved}}{{Images OK}}{{Submitted}}{{Paid}}{{Copyedited}} |
| − | |||
| − | |||
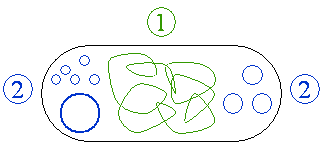
[[image:plasmids.png|frame|right|'''Figure 1''': Schematic drawing of a bacterium with plasmids enclosed. 1 <font color="#339900">Chromosomal DNA</font>. 2 <font color="#0033CC">Plasmids</font>.]] | [[image:plasmids.png|frame|right|'''Figure 1''': Schematic drawing of a bacterium with plasmids enclosed. 1 <font color="#339900">Chromosomal DNA</font>. 2 <font color="#0033CC">Plasmids</font>.]] | ||
| + | A '''plasmid''' is an extrachromosomal, extranuclear [[DNA]] molecule occurring free in the [[cytoplasm]] and capable of autonomous replication (independently of the [[chromosome|chromosomal]] DNA). It usually occurs in [[bacterium|bacteria]], sometimes in [[eukaryote|eukaryotic organisms]] (e.g., the ''2–micron–ring'' in ''Saccharomyces cerevisiae''). | ||
| + | |||
| + | Plasmids are proving to be a great boon for the role of [[human beings]] as stewards of [[nature]]. In addition to research use in [[genetics]] and [[biochemistry]], they serve many practical uses. For example, desirable [[gene]]s can be inserted into plasmids in bacteria, which can then be used to produce large amounts of important proteins, such as [[insulin]] or [[antibiotic]]s, or be used for waste treatment. This level of mastery over creation carries an important responsibility, however, since such human creativity can be used for good (correction of defective genes, treatment of oil spills, production of antibiotics) or for unethical purposes ([[biological warfare]]). | ||
| + | |||
| + | The size of plasmids varies from 1 to over 400 kilobase pairs (kbp). Depending on the size of the plasmids, the number of copies of the same plasmid in a single [[cell (biology)|cell]] varies from one to several hundreds. The larger the plasmid, the fewer is the number of plasmids. The number may even reach thousands for certain artificial plasmids (such as the '''pUC''' series of plasmids) selected for high copy number. | ||
| + | {{toc}} | ||
| + | The term ''plasmid'' was first introduced by the [[United States|American]] [[molecular biology|molecular biologist]] [[Joshua Lederberg]] in 1952 to describe any extrachromosomal hereditary determinant. Currently, however, the term plasmid is restricted only to those accessory DNA molecules that are found in addition to the main chromosomes. | ||
==Characteristic features of Plasmid== | ==Characteristic features of Plasmid== | ||
| − | * | + | * A plasmid typically is a circular and double–stranded [[DNA]] molecule; but linear plasmids have also been reported. |
* It is smaller than and independent of nuclear chromosome. | * It is smaller than and independent of nuclear chromosome. | ||
| − | * It is capable of | + | * It is capable of self–replication using already existing cellular [[enzyme]]s. But the initiation of replication and the distribution of plasmid copies to the daughter cells is controlled by plasmid [[gene]]s. Thus, every plasmid contains at least one DNA sequence that serves as an ''origin of replication,'' or ''ori'' (a starting point for [[DNA replication]]), which enables the plasmid DNA to be duplicated independently from the chromosomal DNA (Figure 4) and to be distributed to the daughter cells. |
| − | * Many plasmids also have genes responsible for phenotypic expression of the organism. | + | * Many plasmids also have genes responsible for [[phenotype|phenotypic]] expression of the organism. |
| − | * Plasmids that exist only as one or a few copies in each bacterium are, upon [[cell division]], in danger of being lost in one of the segregating bacteria. Such | + | * Plasmids that exist only as one or a few copies in each bacterium are, upon [[cell division]], in danger of being lost in one of the segregating bacteria. Such single–copy plasmids have systems that are oriented toward actively distributing a copy to both daughter cells. |
| − | * Some plasmids include an ''addiction system'' or "postsegregational killing system (PSK)" | + | * Some plasmids include an ''addiction system'' or "postsegregational killing system (PSK)." They produce both a long–lived [[poison]] and a short–lived [[antidote]]. Daughter cells that retain a copy of the plasmid survive, while a daughter cell that fails to inherit the plasmid dies or suffers a reduced growth–rate because of the lingering poison from the parent cell. The effect of killing plasmid–free daughter cells appears, at the population level, to ‘‘addict’’ the host to the plasmid. |
| + | |||
| + | ==Important groups of plasmids== | ||
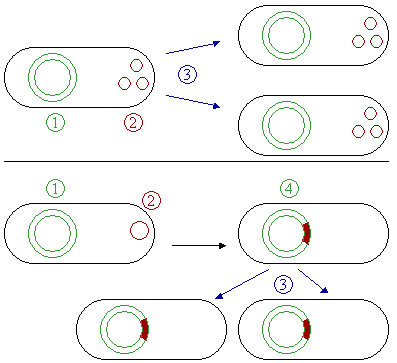
| − | == | + | [[image:plasmid_episome.png|right|frame|'''Figure 2''': Comparison of non–integrating plasmids ''(top)'' and episomes ''(bottom)''. 1 <font color="#009900">Chromosomal DNA</font>. 2 <font color="#990000">Plasmids</font>. 3 <font color="#000099">Cell division</font>. 4 <font color="#009900">Chromosomal DNA</font> with <font color="#990000">integrated plasmids</font>.]] |
| − | [[ | + | Different types of plasmids have been reported and it is possible for plasmids of different varieties to coexist in a single cell. Seven different plasmids have been found in ''[[E. coli]].'' But ''related'' plasmids are often ''incompatible,'' in the sense that only one of them survives in the cell line, due to the regulation of vital plasmid functions. Therefore, plasmids can be assigned into ''compatibility groups.'' Some important groups of plasmids occurring in bacteria are given below. Plasmids can belong to more than one of these functional groups. |
| − | + | ===Episome=== | |
| + | Some plasmids can become associated with another plasmid or with nuclear [[chromosome]]; in the latter case, the plasmid is known as '''episome'''. In another words, an episome is a plasmid of [[bacteria]] or viral DNA that can integrate itself into the chromosomal DNA of the host organism (Figure 2). For this reason, it can stay intact for a long time, be duplicated with every [[cell division]] of the host, and become a basic part of its genetic makeup. | ||
| − | + | Episomes are not essential and may be absent in the cells. They do not originate de novo, but are acquired either due to infection or conjugation. Even if they are present, they may be lost ultimately. This term is no longer commonly used for plasmids, since it is now clear that a transposon (jumping gene or mobile genetic unit) makes a plasmid into an episome. In [[mammal]]ian systems, the term episome refers to a circular DNA (such as a viral genome) that is maintained by non–covalent tethering to the host cell chromosome. | |
| − | |||
| − | |||
| − | === | + | ===F–plasmid=== |
[[Image:BacterConjugation.png|right|350px|thumb|'''Figure 3:''' Overview of Bacterial conjugation]] | [[Image:BacterConjugation.png|right|350px|thumb|'''Figure 3:''' Overview of Bacterial conjugation]] | ||
| − | '''Fertility or | + | '''Fertility or F–plasmids''' are capable of carrying out [[bacterial conjugation]], a plasmid regulated complex process of sexual transfer of F–plasmid to another bacterium (Figure 3). Among the two conjugating strains of bacteria, the one with F–factor is known as male, donor, or F<sup>+</sup> strain and the one without F–factor is known as female, receptor, or F<sup>-</sup> strain. F–factor consists of genes responsible for self–replication, for the formation of ''sex–pili'' or ''f–pili'' to establish contact between the cells of two strains, and for the formation of cytoplasmic bridge and ''tra–genes'' responsible for the transfer of the plasmid. Sometimes, the F–factor can also transfer chromosomal DNA to the receptor bacterium if the factor happens to be integrated to the chromosomal DNA. Such strains have genetic recombination rate 103 times greater than that of F<sup>+</sup> and F<sup>-</sup> strains; so, they are termed ''high frequency recombinants'' (Hfr). Thus, plasmids can be part of the [[mobilome]], since they are often associated with [[conjugation]], a mechanism of [[horizontal gene transfer]]. |
| − | Hence, another way of grouping plasmids is by their ability to transfer genetic material to other bacteria. ''Conjugative'' plasmids contain | + | |
| + | Hence, another way of grouping plasmids is by their ability to transfer genetic material to other bacteria. ''Conjugative'' plasmids contain so–called ''tra–genes,'' which perform the complex process of ''[[Bacterial conjugation|conjugation]].'' ''Non–conjugative'' plasmids are incapable of initiating conjugation; they can only be transferred with the assistance of conjugative plasmids, by "accident." An intermediate class of plasmids carry only a subset of the genes required for transfer and they can "parasitize" a conjugative plasmid, thus transferring at high frequency only in its presence. | ||
| − | === | + | ===R–plasmid=== |
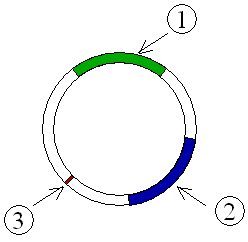
| − | [[image:example_plasmid.png|right|thumb|'''Figure 4''': Schematic drawing of a plasmid with antibiotic resistances. <font color='#009900'>1</font> & <font color='#000099'>2</font> Genes that code for resistance. <font color='#990000'>3</font> Ori.]] | + | [[image:example_plasmid.png|right|thumb|'''Figure 4''': Schematic drawing of a plasmid with [[antibiotic]] resistances. <font color='#009900'>1</font> & <font color='#000099'>2</font> Genes that code for resistance. <font color='#990000'>3</font> Ori.]] |
| − | '''Resistance or | + | '''Resistance or R–plasmids''' contain [[gene]]s or [[gene cassette]]s that confer a selective advantage to the bacterium harboring them, such as the ability to make the bacterium [[antibiotic|antibiotic resistant]], i.e., resistant against antibiotics or [[poison]]s. Historically they were known as R–factors. |
| − | |||
| − | + | Such a plasmid consists of two segments of [[DNA]], one the resistance transfer factor (RTF) responsible for replication as well as transfer of R–plasmid, and the second resistant determinants (r–determinants), which are the [[gene]]s producing substances neutralizing the action of one or another antibiotics or other drugs. These plasmids are ''conjugative'' and spread among the bacteria through conjugation. The antibiotic resistance observed in ''Shigella'' and ''Salmonella'' were due to R–factors. | |
| − | |||
| − | + | One of the early detected R–plasmids is the penicillinase–plasmid of ''Staphylococcus aureus.'' ''S. aureus'' is a [[bacteria|Gram positive]] bacterial pathogen causing infection of skin and wound of humans. After treatment with [[penicillin]] antibiotic, several penicillin–resistant staphylococci developed throughout the world by 1950. High level resistance to penicillin was due to secretion of an [[enzyme]], penicillinase, that degrades penicillin by hydrolysis. | |
| − | |||
| − | === | + | ===Heavy–metal resistance plasmid=== |
| − | + | There are several bacterial strains that contain genetic determinants of resistance to heavy metals, such as Hg<sup>++</sup>, Ag<sup>+</sup>, Cd<sup>++</sup>, CrO<sub>4</sub>, Cu<sup>++</sup>, Ni<sup>++</sup>, Pb<sup>+++</sup>, Zn<sup>++</sup>, and so forth. These determinants for resistance are often found on plasmids and transposons. Bacteria that have been found resistant to heavy metals are ''E. coli,'' ''Pseudomonas aeruginosa,'' ''P. florenscens,'' ''P. syringae,'' and so forth. | |
| − | === | + | ===Col–plasmid=== |
| − | + | There are many bacterial strains that produce [[protein|proteinaceous]] toxins known as ''bacteriocin,'' which are lethal to other strains of the same [[genus]]. The bacteriocin producing genes are present not in [[chromosome]]s, but in special plasmids known as bacteriocin factor. Bacteriocins are already isolated from ''Escherichia coli'' (Colicin), ''Pseudomonas aeruginosa'' (Pyocin), ''Bacillus megaterium'' (Megacine), and others. Toxins secreted by the strains of ''E. coli'' are called [[colicin]]s and the plasmid with responsible gene is known as colicinogeny or Col–factor. Several Col–plasmids like Col B, Col E, Col I, Col V have been recognized and they produce different types of colicins. Some of the Col–plasmids are ''conjugative'' (e.g., Col B, Col V) while others are ''non–conjugative'' (e.g., Col E) and are non–transmissible by their own means. | |
| − | |||
| − | === | + | ===Degradative plasmid=== |
| − | + | '''Degradative plasmids''' consist of genes that equip the bacteria (e.g., ''Pseudomonas sps.'') with special [[enzyme]]s or enzyme system to enable them to digest unusual substances ('''Xenobiotics''') like chlorinated aromatic or hydrocarbon compounds. For example, the camphor (CAM) plasmid of ''P. putida'' encodes enzymes for degradation of camphor, octane (OCT) plasmid helps it degrade octane, XYL–plasmid helps degrade xylene and [[toluene]], NAH–plasmid helps degrade naphthalene, and SAL–plasmid helps it degrade [[salicylic acid|salicilate]]. These plasmids are ''conjugative.'' | |
| − | === | + | ===Virulence plasmid=== |
| − | + | In the race between immunity of the host and virulence of the [[pathogen]], the latter evolves new virulence factors to keep it safely nourished from the host. Much of this virulence is found to be due to the genes in the plasmid known as ''virulence plasmids.'' Formation of invasin due to its virulence plasmid makes ''Shigella flexneri'' (a human intestinal pathogen) able to penetrate intestinal mucosa. Production of '''enterotoxin''' and the cause of diarrhea are also associated with virulence plasmids present in the pathogen. The lysis of R.B.C. ([[erythrocyte]]) of blood caused by some of the pathogenic strains of ''E. coli'' is because of the production of haemolysin from a specific plasmid. | |
| − | + | Another example of virulence plasmid is '''Ti–Plasmid''' (tumor inducing plasmid), which is found in ''Agrobacterium tumefaciens.'' It is a Gram–negative [[soil]] bacterium that infects over 300 different [[dicot]]s, causing crown gall disease at collar region. Ti–plasmid consists of, besides other genes, T–DNA, which encodes enzymes for the synthesis of [[auxin]] and [[cytosine]]. These plant hormones affect plant [[metabolism]], develop tumors, and enable plants to produce compounds called opines, which is used by the pathogen as sources of [[carbon]] and [[nitrogen]]. Closely related to Ti–plasmid is the '''Ri–plasmid''' (root inducing plasmid). It is found in ''A. rhizogenes'' and causes hairy root disease in the plants. | |
| − | |||
| − | + | ===Cryptic plasmid=== | |
| + | During isolation of plasmid DNA from a large number of bacteria, every bacterium was found to contain a low molecular weight DNA as a plasmid. Thus, it seems that the presence of plasmids is a general rule rather than exception. However, not all plasmids consist of genes having any [[phenotype|phenotypic]] significance. The plasmids without any functional genes are termed as cryptic plasmid. | ||
| − | + | ===Vector=== | |
| + | Plasmids used in genetic engineering are called [[vector (biology)|vectors]]. They are used to transfer genes from one organism to another and typically contain a [[genetic marker]] conferring a [[phenotype]] that can be selected for or against. Most of them also contain a [[polylinker]] or multiple cloning site (MCS), which is a short region containing several commonly used restriction sites, allowing the easy insertion of DNA fragments at this location. See [[#Applications|Applications]] below. | ||
| − | + | ==Conformation== | |
| + | DNA can be analyzed by cutting it with restriction enzymes and determining fragment sizes using agarose gel electrophoresis. These enzymes specifically break the DNA at certain short sequences. The resulting linear fragments form "bands" after [[gel electrophoresis]]. It is possible to purify certain fragments by cutting the bands out of the gel and dissolving the gel to release the DNA fragments. | ||
| + | |||
| + | However, uncut plasmid DNA also can be identified. Ultraviolet (UV) treatment can cause dose–dependent incision in the sugar–phosphate backbone of DNA, and the plasmid DNA may appear in one of five conformations, which (for a given size) run at different speeds in a gel during [[electrophoresis]]. The conformations are listed below in order of electrophoretic mobility (speed for a given applied voltage) from slowest to fastest: | ||
| + | * "Nicked Open–Circular" DNA has one strand cut. | ||
| + | * "Linear" DNA has free ends, either because both strands have been cut, or because the DNA was linear ''in vivo.'' You can model this with an electrical extension cord that is not plugged into itself. | ||
| + | * "Relaxed Circular" DNA is fully intact with both strands uncut, but has been enzymatically "relaxed" (supercoils removed). You can model this by letting a twisted extension cord unwind and relax and then plugging it into itself. | ||
| + | * "Supercoiled" (or "Covalently Closed–Circular") DNA is fully intact with both strands uncut, and with a twist built in, resulting in a compact form. You can model this by twisting an extension cord and then plugging it into itself. | ||
| + | * "Supercoiled [[Denaturation (biochemistry)|Denatured]]" DNA is like '''supercoiled DNA''', but has unpaired regions that make it slightly less compact; this can result from excessive alkalinity during plasmid preparation. You can model this by twisting a badly frayed extension cord and then plugging it into itself. | ||
==Plasmid DNA extraction== | ==Plasmid DNA extraction== | ||
| − | For their use as vectors and for | + | For their use as vectors and for molecular cloning, plasmids often need to be isolated. Depending on the need, they can easily be purified away from the rest of the genome. |
| − | There are several methods to isolate plasmid DNA from bacteria, the archetypes of which are the ''' | + | There are several methods to isolate plasmid DNA from bacteria, the archetypes of which are the '''mini–prep''' and the '''maxi–prep'''. The former is a small scale preparation of plasmid DNA and starts with 1-5 ml of an overnight bacterial culture. There are many different miniprep protocols available. Some protocols are variations on the original protocol published by Birnboim and Doly (1979). Others use recent technology and employ a resin that binds, and then elutes, purified plasmid DNA. Several micrograms of DNA can be isolated with the mini–prep, and the procedure can be completed in a relatively short time. Such produced small amounts of impure plasmid DNA is sufficient for analysis by [[restriction digest]] and for some cloning techniques. |
| − | In the latter case, much larger volumes of bacterial suspension are grown from which a | + | In the latter case, much larger volumes (several hundred milliliters) of bacterial suspension are grown from which a maxi–prep can be performed. Formerly, large-scale isolation of plasmid DNA required many hours of ultacentrifugation in a solution of cesium chloride-ethidium bromide. This procedure was time-consuming and required handling of toxic materials. Now, the same resins that can be used for the mini–prep are commonly employed for bulk preparation of the plasmid DNA and the procedure can be completed in several hours. Essentially, this is a scaled–up mini–prep followed by additional purification. This results in relatively large amounts (several μg) of very pure plasmid DNA. |
| − | In recent times many commercial kits have been created to perform plasmid extraction at various scales, purity and levels of automation. Commercial services can prepare plasmid DNA at quoted prices below $ 300 | + | In recent times, many commercial kits have been created to perform plasmid extraction at various scales, purity, and levels of automation. Commercial services can prepare plasmid DNA at quoted prices below US$300 per milligram (mg) in milligram quantities and $15/mg in gram quantities. |
| + | ==Applications== | ||
| + | Plasmids serve as important tools in [[genetics]] and [[biochemistry]] laboratories, where they are commonly used to multiply (make many copies of) or express particular genes. Many plasmids are commercially available for such uses. | ||
| + | For multiplication of genes, the gene to be replicated is inserted into copies of a plasmid that also contains genes that make cells resistant to particular antibiotics. Next, the plasmids are inserted into bacteria by a process called ''[[transformation (genetics)|transformation]].'' Then, the bacteria are exposed to the particular [[antibiotic]]s. Only bacteria which take up copies of the plasmid survive the antibiotic, since the plasmid makes them resistant. In particular, the protecting genes are expressed (used to make a protein) and the expressed protein breaks down the antibiotics. In this way, the antibiotics act as a filter to select only the modified bacteria. These bacteria then can be grown in large amounts, harvested, and [[lyse]]d to isolate the plasmid of interest. | ||
| − | + | Another major use of plasmids is to make large amounts of [[protein]]s. In this case, bacteria are grown that contain a plasmid harboring the gene of interest. Just as the bacteria produce proteins to confer its antibiotic resistance; it can also be induced to produce large amounts of proteins from the inserted gene. This is a cheap and easy way of mass–producing a gene or the protein it then codes for, for example, [[insulin]] or even antibiotics. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | Desired genes can be inserted in the plasmid and bacteria can be made to express the phenotype of the gene, and through genetic engineering one can design bacteria for decomposition of xenobiotics, waste water treatment, and many other works. Moreover, plasmids can be good tools for introducing supplementary copies of defective gene to cure hereditary [[disease]]s in both [[human]] as well as in [[animal]]s. | |
| − | == | + | ==References== |
| − | * [[ | + | * Bimboim, H. C. and J. Doly. 1979. [http://nar.oxfordjournals.org/cgi/content/abstract/7/6/1513 A rapid alkaline extraction procedure for screening recombinant plasmid DNA] ''Nucl. Acids Res.'' 7: 1513-1523. Retrieved May 23, 2007. |
| + | * Cooper, T. F., and J. A. Heinemann. 2000. [http://www.pnas.org/cgi/reprint/97/23/12643.pdf Postsegregational killing does not increase plasmid stability but acts to mediate the exclusion of competing plasmids]. ''PNAS'' 97: 12643-12648. Retrieved May 22, 2007. | ||
| + | * Lin–Chao, S., W. T. Chen, and T. T. Wong. 1992. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=1283002 High copy number of the pUC plasmid results from a Rom/Rop-suppressible point mutation in RNA II]. ''Mol. Microbiol.'' 6(22): 3385-3393. Retrieved May 22, 2007. | ||
| + | * Pelczar, M. J., E. C. S. Chan, and N. R. Krieg. 1993. ''Microbiology: Concepts and Applications.'' McGraw–Hill, Inc., New York. ISBN 0070492581. | ||
| + | * Schlegel, H. G., and C. Zaborosch. 1992. ''Allgemeine Mikrobiologie.'' Georg Thieme Verlag Stuttgart. ISBN 3134446073. | ||
| + | * Sheua, S.–Y., W.–M. Chenb, and G.–H. Linc. 2007. [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WPF-4MP5KSH-1&_user=10&_coverDate=05%2F31%2F2007&_rdoc=4&_fmt=summary&_orig=browse&_srch=doc-info(%23toc%236989%232007%23999429996%23650221%23FLA%23display%23Volume)&_cdi=6989&_sort=d&_docanchor=&_ct=9&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=22618fb12bc436fc79b99a15998745C.E. Characterization and application of a rolling-circle-type plasmid from Cupriavidus taiwanensis]. ''Plasmid'' 57(3): 275-285. Retrieved May 22, 2007. | ||
| + | * Veit, B. E., and W. L. Fangman. 1988. [http://www.pubmedcentral.nih.gov/pagerender.fcgi?artid=365588&pageindex=1 Copy number and partition of the Saccharomyces cerevisiae 2 micron plasmid controlled by transcription regulators]. ''Mol. Cell Biol.'' 8(11): 4949–4957. Retrieved May 22, 2007. | ||
| − | |||
| − | |||
| − | |||
| − | |||
{{credit|113504485}} | {{credit|113504485}} | ||
[[Category:Life sciences]] | [[Category:Life sciences]] | ||
| + | [[Category:Molecular biology]] | ||
| + | [[Category:Microbiology]] | ||
| + | [[Category:Cell biology]] | ||
Latest revision as of 19:27, 6 May 2015
A plasmid is an extrachromosomal, extranuclear DNA molecule occurring free in the cytoplasm and capable of autonomous replication (independently of the chromosomal DNA). It usually occurs in bacteria, sometimes in eukaryotic organisms (e.g., the 2–micron–ring in Saccharomyces cerevisiae).
Plasmids are proving to be a great boon for the role of human beings as stewards of nature. In addition to research use in genetics and biochemistry, they serve many practical uses. For example, desirable genes can be inserted into plasmids in bacteria, which can then be used to produce large amounts of important proteins, such as insulin or antibiotics, or be used for waste treatment. This level of mastery over creation carries an important responsibility, however, since such human creativity can be used for good (correction of defective genes, treatment of oil spills, production of antibiotics) or for unethical purposes (biological warfare).
The size of plasmids varies from 1 to over 400 kilobase pairs (kbp). Depending on the size of the plasmids, the number of copies of the same plasmid in a single cell varies from one to several hundreds. The larger the plasmid, the fewer is the number of plasmids. The number may even reach thousands for certain artificial plasmids (such as the pUC series of plasmids) selected for high copy number.
The term plasmid was first introduced by the American molecular biologist Joshua Lederberg in 1952 to describe any extrachromosomal hereditary determinant. Currently, however, the term plasmid is restricted only to those accessory DNA molecules that are found in addition to the main chromosomes.
Characteristic features of Plasmid
- A plasmid typically is a circular and double–stranded DNA molecule; but linear plasmids have also been reported.
- It is smaller than and independent of nuclear chromosome.
- It is capable of self–replication using already existing cellular enzymes. But the initiation of replication and the distribution of plasmid copies to the daughter cells is controlled by plasmid genes. Thus, every plasmid contains at least one DNA sequence that serves as an origin of replication, or ori (a starting point for DNA replication), which enables the plasmid DNA to be duplicated independently from the chromosomal DNA (Figure 4) and to be distributed to the daughter cells.
- Many plasmids also have genes responsible for phenotypic expression of the organism.
- Plasmids that exist only as one or a few copies in each bacterium are, upon cell division, in danger of being lost in one of the segregating bacteria. Such single–copy plasmids have systems that are oriented toward actively distributing a copy to both daughter cells.
- Some plasmids include an addiction system or "postsegregational killing system (PSK)." They produce both a long–lived poison and a short–lived antidote. Daughter cells that retain a copy of the plasmid survive, while a daughter cell that fails to inherit the plasmid dies or suffers a reduced growth–rate because of the lingering poison from the parent cell. The effect of killing plasmid–free daughter cells appears, at the population level, to ‘‘addict’’ the host to the plasmid.
Important groups of plasmids
Different types of plasmids have been reported and it is possible for plasmids of different varieties to coexist in a single cell. Seven different plasmids have been found in E. coli. But related plasmids are often incompatible, in the sense that only one of them survives in the cell line, due to the regulation of vital plasmid functions. Therefore, plasmids can be assigned into compatibility groups. Some important groups of plasmids occurring in bacteria are given below. Plasmids can belong to more than one of these functional groups.
Episome
Some plasmids can become associated with another plasmid or with nuclear chromosome; in the latter case, the plasmid is known as episome. In another words, an episome is a plasmid of bacteria or viral DNA that can integrate itself into the chromosomal DNA of the host organism (Figure 2). For this reason, it can stay intact for a long time, be duplicated with every cell division of the host, and become a basic part of its genetic makeup.
Episomes are not essential and may be absent in the cells. They do not originate de novo, but are acquired either due to infection or conjugation. Even if they are present, they may be lost ultimately. This term is no longer commonly used for plasmids, since it is now clear that a transposon (jumping gene or mobile genetic unit) makes a plasmid into an episome. In mammalian systems, the term episome refers to a circular DNA (such as a viral genome) that is maintained by non–covalent tethering to the host cell chromosome.
F–plasmid
Fertility or F–plasmids are capable of carrying out bacterial conjugation, a plasmid regulated complex process of sexual transfer of F–plasmid to another bacterium (Figure 3). Among the two conjugating strains of bacteria, the one with F–factor is known as male, donor, or F+ strain and the one without F–factor is known as female, receptor, or F- strain. F–factor consists of genes responsible for self–replication, for the formation of sex–pili or f–pili to establish contact between the cells of two strains, and for the formation of cytoplasmic bridge and tra–genes responsible for the transfer of the plasmid. Sometimes, the F–factor can also transfer chromosomal DNA to the receptor bacterium if the factor happens to be integrated to the chromosomal DNA. Such strains have genetic recombination rate 103 times greater than that of F+ and F- strains; so, they are termed high frequency recombinants (Hfr). Thus, plasmids can be part of the mobilome, since they are often associated with conjugation, a mechanism of horizontal gene transfer.
Hence, another way of grouping plasmids is by their ability to transfer genetic material to other bacteria. Conjugative plasmids contain so–called tra–genes, which perform the complex process of conjugation. Non–conjugative plasmids are incapable of initiating conjugation; they can only be transferred with the assistance of conjugative plasmids, by "accident." An intermediate class of plasmids carry only a subset of the genes required for transfer and they can "parasitize" a conjugative plasmid, thus transferring at high frequency only in its presence.
R–plasmid
Resistance or R–plasmids contain genes or gene cassettes that confer a selective advantage to the bacterium harboring them, such as the ability to make the bacterium antibiotic resistant, i.e., resistant against antibiotics or poisons. Historically they were known as R–factors.
Such a plasmid consists of two segments of DNA, one the resistance transfer factor (RTF) responsible for replication as well as transfer of R–plasmid, and the second resistant determinants (r–determinants), which are the genes producing substances neutralizing the action of one or another antibiotics or other drugs. These plasmids are conjugative and spread among the bacteria through conjugation. The antibiotic resistance observed in Shigella and Salmonella were due to R–factors.
One of the early detected R–plasmids is the penicillinase–plasmid of Staphylococcus aureus. S. aureus is a Gram positive bacterial pathogen causing infection of skin and wound of humans. After treatment with penicillin antibiotic, several penicillin–resistant staphylococci developed throughout the world by 1950. High level resistance to penicillin was due to secretion of an enzyme, penicillinase, that degrades penicillin by hydrolysis.
Heavy–metal resistance plasmid
There are several bacterial strains that contain genetic determinants of resistance to heavy metals, such as Hg++, Ag+, Cd++, CrO4, Cu++, Ni++, Pb+++, Zn++, and so forth. These determinants for resistance are often found on plasmids and transposons. Bacteria that have been found resistant to heavy metals are E. coli, Pseudomonas aeruginosa, P. florenscens, P. syringae, and so forth.
Col–plasmid
There are many bacterial strains that produce proteinaceous toxins known as bacteriocin, which are lethal to other strains of the same genus. The bacteriocin producing genes are present not in chromosomes, but in special plasmids known as bacteriocin factor. Bacteriocins are already isolated from Escherichia coli (Colicin), Pseudomonas aeruginosa (Pyocin), Bacillus megaterium (Megacine), and others. Toxins secreted by the strains of E. coli are called colicins and the plasmid with responsible gene is known as colicinogeny or Col–factor. Several Col–plasmids like Col B, Col E, Col I, Col V have been recognized and they produce different types of colicins. Some of the Col–plasmids are conjugative (e.g., Col B, Col V) while others are non–conjugative (e.g., Col E) and are non–transmissible by their own means.
Degradative plasmid
Degradative plasmids consist of genes that equip the bacteria (e.g., Pseudomonas sps.) with special enzymes or enzyme system to enable them to digest unusual substances (Xenobiotics) like chlorinated aromatic or hydrocarbon compounds. For example, the camphor (CAM) plasmid of P. putida encodes enzymes for degradation of camphor, octane (OCT) plasmid helps it degrade octane, XYL–plasmid helps degrade xylene and toluene, NAH–plasmid helps degrade naphthalene, and SAL–plasmid helps it degrade salicilate. These plasmids are conjugative.
Virulence plasmid
In the race between immunity of the host and virulence of the pathogen, the latter evolves new virulence factors to keep it safely nourished from the host. Much of this virulence is found to be due to the genes in the plasmid known as virulence plasmids. Formation of invasin due to its virulence plasmid makes Shigella flexneri (a human intestinal pathogen) able to penetrate intestinal mucosa. Production of enterotoxin and the cause of diarrhea are also associated with virulence plasmids present in the pathogen. The lysis of R.B.C. (erythrocyte) of blood caused by some of the pathogenic strains of E. coli is because of the production of haemolysin from a specific plasmid.
Another example of virulence plasmid is Ti–Plasmid (tumor inducing plasmid), which is found in Agrobacterium tumefaciens. It is a Gram–negative soil bacterium that infects over 300 different dicots, causing crown gall disease at collar region. Ti–plasmid consists of, besides other genes, T–DNA, which encodes enzymes for the synthesis of auxin and cytosine. These plant hormones affect plant metabolism, develop tumors, and enable plants to produce compounds called opines, which is used by the pathogen as sources of carbon and nitrogen. Closely related to Ti–plasmid is the Ri–plasmid (root inducing plasmid). It is found in A. rhizogenes and causes hairy root disease in the plants.
Cryptic plasmid
During isolation of plasmid DNA from a large number of bacteria, every bacterium was found to contain a low molecular weight DNA as a plasmid. Thus, it seems that the presence of plasmids is a general rule rather than exception. However, not all plasmids consist of genes having any phenotypic significance. The plasmids without any functional genes are termed as cryptic plasmid.
Vector
Plasmids used in genetic engineering are called vectors. They are used to transfer genes from one organism to another and typically contain a genetic marker conferring a phenotype that can be selected for or against. Most of them also contain a polylinker or multiple cloning site (MCS), which is a short region containing several commonly used restriction sites, allowing the easy insertion of DNA fragments at this location. See Applications below.
Conformation
DNA can be analyzed by cutting it with restriction enzymes and determining fragment sizes using agarose gel electrophoresis. These enzymes specifically break the DNA at certain short sequences. The resulting linear fragments form "bands" after gel electrophoresis. It is possible to purify certain fragments by cutting the bands out of the gel and dissolving the gel to release the DNA fragments.
However, uncut plasmid DNA also can be identified. Ultraviolet (UV) treatment can cause dose–dependent incision in the sugar–phosphate backbone of DNA, and the plasmid DNA may appear in one of five conformations, which (for a given size) run at different speeds in a gel during electrophoresis. The conformations are listed below in order of electrophoretic mobility (speed for a given applied voltage) from slowest to fastest:
- "Nicked Open–Circular" DNA has one strand cut.
- "Linear" DNA has free ends, either because both strands have been cut, or because the DNA was linear in vivo. You can model this with an electrical extension cord that is not plugged into itself.
- "Relaxed Circular" DNA is fully intact with both strands uncut, but has been enzymatically "relaxed" (supercoils removed). You can model this by letting a twisted extension cord unwind and relax and then plugging it into itself.
- "Supercoiled" (or "Covalently Closed–Circular") DNA is fully intact with both strands uncut, and with a twist built in, resulting in a compact form. You can model this by twisting an extension cord and then plugging it into itself.
- "Supercoiled Denatured" DNA is like supercoiled DNA, but has unpaired regions that make it slightly less compact; this can result from excessive alkalinity during plasmid preparation. You can model this by twisting a badly frayed extension cord and then plugging it into itself.
Plasmid DNA extraction
For their use as vectors and for molecular cloning, plasmids often need to be isolated. Depending on the need, they can easily be purified away from the rest of the genome.
There are several methods to isolate plasmid DNA from bacteria, the archetypes of which are the mini–prep and the maxi–prep. The former is a small scale preparation of plasmid DNA and starts with 1-5 ml of an overnight bacterial culture. There are many different miniprep protocols available. Some protocols are variations on the original protocol published by Birnboim and Doly (1979). Others use recent technology and employ a resin that binds, and then elutes, purified plasmid DNA. Several micrograms of DNA can be isolated with the mini–prep, and the procedure can be completed in a relatively short time. Such produced small amounts of impure plasmid DNA is sufficient for analysis by restriction digest and for some cloning techniques.
In the latter case, much larger volumes (several hundred milliliters) of bacterial suspension are grown from which a maxi–prep can be performed. Formerly, large-scale isolation of plasmid DNA required many hours of ultacentrifugation in a solution of cesium chloride-ethidium bromide. This procedure was time-consuming and required handling of toxic materials. Now, the same resins that can be used for the mini–prep are commonly employed for bulk preparation of the plasmid DNA and the procedure can be completed in several hours. Essentially, this is a scaled–up mini–prep followed by additional purification. This results in relatively large amounts (several μg) of very pure plasmid DNA.
In recent times, many commercial kits have been created to perform plasmid extraction at various scales, purity, and levels of automation. Commercial services can prepare plasmid DNA at quoted prices below US$300 per milligram (mg) in milligram quantities and $15/mg in gram quantities.
Applications
Plasmids serve as important tools in genetics and biochemistry laboratories, where they are commonly used to multiply (make many copies of) or express particular genes. Many plasmids are commercially available for such uses.
For multiplication of genes, the gene to be replicated is inserted into copies of a plasmid that also contains genes that make cells resistant to particular antibiotics. Next, the plasmids are inserted into bacteria by a process called transformation. Then, the bacteria are exposed to the particular antibiotics. Only bacteria which take up copies of the plasmid survive the antibiotic, since the plasmid makes them resistant. In particular, the protecting genes are expressed (used to make a protein) and the expressed protein breaks down the antibiotics. In this way, the antibiotics act as a filter to select only the modified bacteria. These bacteria then can be grown in large amounts, harvested, and lysed to isolate the plasmid of interest.
Another major use of plasmids is to make large amounts of proteins. In this case, bacteria are grown that contain a plasmid harboring the gene of interest. Just as the bacteria produce proteins to confer its antibiotic resistance; it can also be induced to produce large amounts of proteins from the inserted gene. This is a cheap and easy way of mass–producing a gene or the protein it then codes for, for example, insulin or even antibiotics.
Desired genes can be inserted in the plasmid and bacteria can be made to express the phenotype of the gene, and through genetic engineering one can design bacteria for decomposition of xenobiotics, waste water treatment, and many other works. Moreover, plasmids can be good tools for introducing supplementary copies of defective gene to cure hereditary diseases in both human as well as in animals.
ReferencesISBN links support NWE through referral fees
- Bimboim, H. C. and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA Nucl. Acids Res. 7: 1513-1523. Retrieved May 23, 2007.
- Cooper, T. F., and J. A. Heinemann. 2000. Postsegregational killing does not increase plasmid stability but acts to mediate the exclusion of competing plasmids. PNAS 97: 12643-12648. Retrieved May 22, 2007.
- Lin–Chao, S., W. T. Chen, and T. T. Wong. 1992. High copy number of the pUC plasmid results from a Rom/Rop-suppressible point mutation in RNA II. Mol. Microbiol. 6(22): 3385-3393. Retrieved May 22, 2007.
- Pelczar, M. J., E. C. S. Chan, and N. R. Krieg. 1993. Microbiology: Concepts and Applications. McGraw–Hill, Inc., New York. ISBN 0070492581.
- Schlegel, H. G., and C. Zaborosch. 1992. Allgemeine Mikrobiologie. Georg Thieme Verlag Stuttgart. ISBN 3134446073.
- Sheua, S.–Y., W.–M. Chenb, and G.–H. Linc. 2007. Characterization and application of a rolling-circle-type plasmid from Cupriavidus taiwanensis. Plasmid 57(3): 275-285. Retrieved May 22, 2007.
- Veit, B. E., and W. L. Fangman. 1988. Copy number and partition of the Saccharomyces cerevisiae 2 micron plasmid controlled by transcription regulators. Mol. Cell Biol. 8(11): 4949–4957. Retrieved May 22, 2007.
Credits
New World Encyclopedia writers and editors rewrote and completed the Wikipedia article in accordance with New World Encyclopedia standards. This article abides by terms of the Creative Commons CC-by-sa 3.0 License (CC-by-sa), which may be used and disseminated with proper attribution. Credit is due under the terms of this license that can reference both the New World Encyclopedia contributors and the selfless volunteer contributors of the Wikimedia Foundation. To cite this article click here for a list of acceptable citing formats.The history of earlier contributions by wikipedians is accessible to researchers here:
The history of this article since it was imported to New World Encyclopedia:
Note: Some restrictions may apply to use of individual images which are separately licensed.