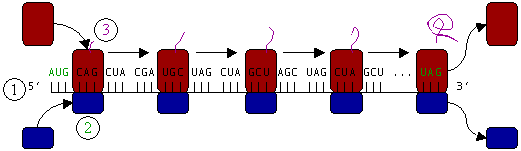Difference between revisions of "Ribosome" - New World Encyclopedia
({{Contracted}}) |
|||
| (23 intermediate revisions by 8 users not shown) | |||
| Line 1: | Line 1: | ||
| − | {{ | + | {{Ebapproved}}{{Approved}}{{Images OK}}{{Submitted}}{{Paid}}{{copyedited}}{{2Copyedited}} |
| − | + | A '''ribosome''' is a small, dense granular particle comprising usually three or four [[rRNA|ribosomal RNA]] molecules and more than 50 [[protein]] molecules, interconnected to form the site of protein synthesis. The ribosome is the site at which the [[mRNA|messenger RNA]]'s code for linking amino acids together in a chain to form a particular new protein is translated into that protein or polypeptide. Ribosomes occur in both [[prokaryote|prokaryotic]] and [[eukaryote|eukaryotic]] [[cell (biology)|cell]]s. Ribosomes from [[bacteria]] and [[archaea]] are smaller than the ribosomes from [[eukaryote]]s, although all three domains of life have significantly different ribosomes. Interestingly, the ribosomes in the [[mitochondrion]] of eukaryotic cells resemble those in bacteria, reflecting the assumed evolutionary origin of this [[organelle]] (Benne and Sloof 1987). | |
| − | A | + | A ribosome can be thought of as a giant [[enzyme]] that builds proteins from a set of genetic instructions. The ribosome's enzymatic activity derives from the presence of the rRNA, which in acting as an enzyme exemplifies a [[ribozyme]] and lends credence to the RNA world hypothesis that in the origins of life, RNA preceded DNA. Ribosomes occur either freely, as in the matrix of [[mitochondria]], [[chloroplast]]s, and [[cytoplasm]] (the internal fluid of the cell), or in a membrane-bound state, as in the [[endoplasmic reticulum]] and the [[nuclear envelope]]. |
| + | {{toc}} | ||
| + | The intricate process by which messenger RNA is translated into a specific sequence of amino acids is a testimony to the remarkable complexity and harmony in the universe. The process has to be very precise, otherwise the functionality of the protein could be compromised. | ||
| − | Ribosomes | + | ==Overview== |
| − | + | ||
| + | Ribosomes, the sites of [[protein]] synthesis within living cells, receive instructions from the DNA genes through messenger ribonucleic acid (mRNA), encoding a chemical "blueprint" for a protein product. A protein is a complex, high-molecular mass organic compound comprising [[amino acid]]s joined together in chains, called peptides or polypeptides depending on their length. Transfer RNA (tRNA) is a small RNA chain (73-93 nucleotides) that transfers a specific amino acid to a growing polypeptide chain at the ribosomal site of protein synthesis during translation of the mRNA into a protein. | ||
| + | |||
| + | The rRNA portions of the ribosome perform the catalytic processes by which ribosomes synthesize proteins while the protein portions of the ribosome support the function of the rRNA [[ribozyme]] (an RNA molecule that catalyzes a chemical reaction). Such evidence lends support to the RNA world hypothesis, which proposes that a world filled with RNA (ribonucleic acid) based life predates current world filled with DNA (deoxyribonucleic acid) based life. In the proposed RNA world, RNA would have served some of the catalytic functions now served by proteins, and the ribosomes would be a remnant from that world now deposed by the DNA world. | ||
| + | |||
| + | Ribosomes were first clearly described by [[Romania|Romanian]] cell biologist [[George Palade]] in the mid–1950s, as dense particles or granules of ribonucleoprotein, after he observed them under the [[electron microscope]] (Palade 1955). For this, Palade would win the [[Nobel Prize]]. The term "ribosome" was later proposed by the scientist Richard B. Roberts in 1958, while writing the introductory comments for the symposium proceedings "Microsomal Particles and Protein Synthesis" (Roberts 1958). | ||
| + | |||
| + | The structure and function of the ribosomes and associated molecules, known as the ''translational apparatus,'' has been of research interest since the mid-20<sup>th</sup> century and the focus of the study has been to work out the topology (shape and positions of the individual protein and rRNA) of ribosomes. | ||
| + | |||
| + | ==Occurrence== | ||
| − | + | Ribosomes are abundant components of both prokaryotic and eukaryotic cells and of both [[plant]] and [[animal]] cells. An ''Escherichia coli'' cell contains roughly 10,000 ribosomes, which together form about 25 percent of the total [[bacteria]]l cell mass. A [[mammal]]ian cell may contain as many as 10 million ribosomes. In [[prokaryote|prokaryotic]] cells, the ribosomes are distributed freely in the [[cytoplasm]]. In [[eukaryote|eukaryotic]] cells, they are found either freely floating in the matrix of [[mitochondria]], [[chloroplast]]s, and cytoplasm or attached to the membrane of the [[endoplasmic reticulum]] and the nuclear envelope. | |
| − | + | Free and membrane–bound ribosomes differ only in their spatial distribution; they are identical in structure and function. Whether the ribosome exists in a free or membrane–bound state depends on the presence of an [[signal peptide|ER targeting signal sequence]] on the protein being synthesized. | |
| − | |||
| − | |||
| − | |||
===Free ribosomes=== | ===Free ribosomes=== | ||
| − | |||
| − | + | Free ribosomes are "free" to move about anywhere in the cytoplasm (within the [[cell membrane]]). [[Yeast]] cells, [[reticulocytes]] or [[lymphocyte]]s, [[meristem|meristematic]] plant tissues, [[embryo|embryonic]] nerve cells, and [[cancer|cancerous]] cells contain a large number of free ribosomes. Proteins made by free ribosomes are used within the cell. Thus, the cells that synthesize specific proteins for the intracellular utilization and storage often contain large number of free ribosomes. Such cells include [[erythroblast]]s, developing [[muscle]] cells, [[skin]] cells, and so forth. | |
| − | + | ||
| + | ===Membrane–bound ribosomes=== | ||
| − | + | When certain proteins are synthesized, they need be "membrane–bound." Therefore, the new polypeptide chains are usually synthesized in membrane-bound ribosomes and are inserted directly into the [[endoplasmic reticulum]], from where they are then transported to their destinations. Bound ribosomes usually produce proteins that are used within the cell membrane or are expelled from the cell via ''[[exocytosis]]''. Thus, in the cells actively engaged in protein synthesis, the ribosomes tend to remain attached to the membranes of the endoplasmic reticulum. Such cells include the [[pancreas|pancreatic]] cells, hepatic parenchymal cells, [[osteoblast]]s, serous cells, or submaxillary gland cells, [[mammary gland]] cells, [[thyroid]] cells, and the chief cells of the glandular [[stomach]] in [[bird]]s, [[rodent]]s, and [[ruminant]]s. | |
==Structure== | ==Structure== | ||
| − | |||
| − | Prokaryotes have 70[[Svedberg | + | ===Overview=== |
| + | |||
| + | The various ribosomes share a core structure that is quite similar despite the large differences in size. Ribosomes are oblate spheroid granules with a diameter ranging from 15 to 25 nanometers (150 to 250 Å). Each ribosome is porous, hydrated, and consists of two subunits (Figure 1). One ribosomal subunit is larger in size and has a dome–like shape, while the other ribosomal subunit is smaller and occurs above the larger one forming a cap–like structure (Figure 2). The ribosomes are chemically composed mainly of [[RNA]] (ribosomal RNA, rRNA) and [[protein]]s and thus are called ribonucleoproteins, RNPs. Both constituents occur approximately in equal proportion in its two subunits. [[Lipid]] is totally absent in ribosomes. | ||
| + | |||
| + | [[Crystallography|Crystallographic]] work has shown that there are no ribosomal proteins close to the reaction site for polypeptide synthesis, which is fully occupied by the rRNA. This suggests that the ribosomal protein does not participate directly in the synthesis of proteins, but rather provides a scaffold that enhances the ability of rRNA to synthesize protein (Alberts et al. 2002). | ||
| + | |||
| + | The two ribosomal subunits remain fit together due to a high concentration of Mg<sup>++</sup> ions. In a decreased Mg<sup>++</sup> concentration, the two subunits dissociate. Actually, in bacterial cells, the two subunits are found to occur freely in the cytoplasm and they come together only for the process of protein synthesis. At high concentration of Mg<sup>++</sup> ions in the matrix, two ribosomes (each called '''monosomes''') become associated with each other and form what is known as '''dimer.''' Further, during the process of protein synthesis, several ribosomes are often working their way along the same mRNA, somewhat like beads on a string; the resulting structure is known as '''polyribosome''' or '''polysome.''' | ||
| + | [[Image:ribosome_structure_fixed.png|frame|'''Figure 2: ''' Large (1) and small (2) subunit fit together]] | ||
| + | |||
| + | ===Prokaryotic ribosomes=== | ||
| + | Prokaryotes have comparatively smaller ribosomes with the sedimentation coefficient of 70 [[Svedberg]] units (abbreviated as '''S'''), and a molecular weight of 2.7x10<sup>6</sup> [[dalton]]s. Each of the 70S ribosomes comprises a small (30S) and a large (50S) subunit. (Svedberg measures are not additive because sedimentation rate depends on both mass and surface area.) The 70S ribosomes contain proportionally more [[RNA]] than protein. For example, the ribosomes of ''E. coli'' contain 63 percent rRNA and 37 percent protein. The 70S ribosomes have three different types of rRNA: '''23S rRNA,''' '''16S rRNA,''' and '''5S rRNA.''' The large subunit is composed of a 5S rRNA subunit (comprising 120 nucleotides), a 23S rRNA subunit (comprising 2900 nucleotides), and 34 [[protein]]s. The 30S subunit has a 16S rRNA subunit (comprising 1540 nucleotides) bound to 21 proteins (Alberts et al. 2002). | ||
| + | |||
| + | ===Eukaryotic ribosomes=== | ||
| + | Eukaryotes have bigger ribosomes of 80S sedimentation coefficient and of 40x10<sup>6</sup> daltons molecular weight. Each 80S ribosome comprises a small (40S) and a large (60S) subunit. The ribosomal subunits of [[prokaryote]]s and [[eukaryote]]s are quite similar (Alberts et al. 2002). However, 80S ribosomes are composed of proportionally less RNA and more protein. For example, in pea seedling, ribosomes comprise 40 percent rRNA and 60 percent protein. | ||
| + | |||
| + | There are four different types of rRNA in 80S ribosomes: '''28S rRNA''' (but '''25–26S rRNA''' in [[plant]]s, [[fungi]], and [[protozoan]]s), '''18S rRNA,''' '''5S rRNA,''' and '''5.8S rRNA.''' The large 60S subunit comprises a 5S RNA (120 nucleotides), a 28S RNA (4700 nucleotides), and a 5.8S RNA (160 nucleotides) subunit, and about 49 proteins. The 40S subunit comprises an 18S RNA (1900 nucleotides) subunit and about 33 proteins (Alberts et al. 2002). About 60 percent of the rRNA is helical (i.e., double stranded) and contains paired bases. These double stranded regions are due to hairpin loops between complimentary regions of the linear molecule. Thus, it can be said that the extra [[RNA]] in the larger ribosomes is in several long continuous insertions, such that they form loops out of the core structure without disrupting or changing it (Alberts et al. 2002). | ||
| − | + | The ribosomes found in [[chloroplast]]s and [[mitochondria]] of eukaryotes also comprise large and small subunits bound together into one 55S particle (Alberts et al. 2002). These [[organelle]]s are believed to be descendants of [[bacteria]] (see [[Endosymbiotic theory]]) and as such their ribosomes are similar to those of prokaryotes (Alberts et al. 2002). The 55S ribosomes of mammalian mitochondria lack 5S rRNA, but contain 21S and 12S rRNAs. The 21S rRNA occurs in larger or 35S ribosomal subunit, while 12S rRNA occurs in smaller or 25S ribosomal subunit. | |
| − | The ribosomes | + | The differences between the prokaryotic and eukaryotic ribosomes are exploited by [[medicinal chemistry|pharmaceutical chemists]] to create [[antibiotic]]s that can destroy a bacterial infection without harming the cells of the infected person. Due to differences in their structures, the bacterial 70S ribosomes are vulnerable to these antibiotics (such as Chloramphenicol) while the eukaryotic 80S ribosomes are not. Even though [[mitochondria]] possess ribosomes similar to the bacterial ones, mitochondria are not affected by these antibiotics because they are surrounded by a double membrane that does not easily admit these antibiotics into the [[organelle]] (O'Brien 1971). |
| − | + | ===Ultra–structure=== | |
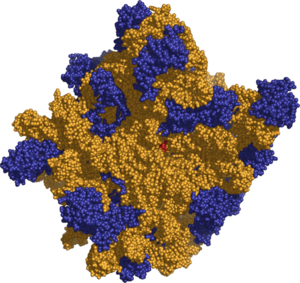
| + | [[Image:Ribosome 50s.png|thumb|300px|right|'''Figure 3 :''' Molecular structure of the 50S Subunit. Proteins are shown in blue and RNA in orange.]] | ||
| + | The general molecular structure of the ribosome has been known since the early 1970s. In the early 2000s, the structure was revealed at high resolutions, in the order of a few angstroms, and scientific papers presenting the structure of the ribosome at atomic resolution were published in rapid succession in late 2000. | ||
| − | The | + | The structure of the 30S small subunit from ''Thermus thermophilus'' (a highly [[thermophilic]] [[bacteria]] first discovered in deep–sea hot vents) shows that the decoding center, which positions mRNA and three tRNAs, is constructed entirely of RNA (Schluenzen et al., 2000; Wimberly, 2000). The mRNA threads through a tunnel within the small subunit. The 3’ end of the 16S rRNA is supposed to be involved in mRNA binding. Each of the three tRNAs is bound in distinctive binding sites made from structural elements contributed by both the 50S subunit and the 30S subunit. In each of the three tRNA binding sites (A–, P–, and E–sites), the ribosome contacts all of the major elements of tRNA, providing an explanation for the conservation of tRNA structure (Yusupov et al. 2001). The anticodon stem–loops of tRNAs point into the 30S subunit, whereas 3’ ends attached to amino acid or peptide through an acyl bond point down in 50S subunit. Further, the 3’ ends of the A–site and P–site tRNAs are juxtaposed in the peptidyl transferase site of the 50S subunit. However, a metal ion stabilizes a kink in the mRNA that demarcates the boundary between A and P sites, which is potentially important to prevent slippage of mRNA, while translocation of tRNA from site A to P. Also, metal ions stabilize the inter–subunit interface (Selmer 2006). |
| − | + | The 16S rRNA of the small, 30S subunit folds into four domains: 5', central, 3' major, and 3' minor. The structural autonomy of these domains implies that they move relative to one another. Thus, the head of the small subunit shows a high degree of flexibility compared to its rest of the body. Swiveling of the head observed by Schuwirth et al. (2005) suggests a mechanism for the final movements of messenger RNA (mRNA) and transfer RNAs (tRNAs) during translocation. Structural changes correlating events at the particle's far end with the cycle of mRNA translocation at the decoding region are transmitted by extended RNA helical elements that run longitudinally through its body (Schluenzen et al. 2000). | |
| − | |||
| − | The | ||
| − | The | + | The 23S rRNA of the large, 50S subunit folds into six secondary structural domains, while the seventh domain is formed by 5S rRNA. The 50S subunit from the archea, ''Haloarcula marismortui'' is shown to have all its rRNAs fit together like the pieces of a three–dimensional jigsaw puzzle to form a large, monolithic structure. Proteins are found everywhere on its surface except in the active site where [[peptide]] bond formation takes place and where it contacts the small subunit. Most of the proteins stabilize the structure by interacting with several rRNA domains (Ban et al. 2000). The large ribosomal subunit catalyzes peptide bond formation and binds factors for initiation, termination, and elongation. The peptidyl transferase function is attributed to the 23S rRNA, making this RNA a "ribozyme." Nascent polypeptides emerge through a tunnel in the large ribosome subunit. The tunnel lumen is lined with RNA helices and some ribosomal protein. |
| − | + | ==Biogenesis== | |
| + | Ribosomes are not self–replicating particles. Synthesis of various components of ribosomes such as rRNAs and proteins is under genetic control. In bacteria, a single gene transcript containing the sequences for 16S, 23S, and 5S rRNAs is synthesized by an rRNA operon (transcriptional unit for multiple molecules) and this larger molecule undergoes both tailoring and chemical modifications before each rRNA molecule assumes its mature form. The whole process of biosynthesis of 70S ribosomes takes place in the cytoplasm. The required amount of ribonucleoprotein synthesis is under autogenous regulation of translation. | ||
| − | + | In eukaryotes, the biogenesis of ribosomes is much more complex involving three main events: rRNA synthesis by [[nucleolus|nucleolar]] organizer (NO), synthesis of 5S rRNA, and biosynthesis of ribonucleoprotein. The 5.8S, 18S, and 28S rRNAs are transcribed as a much larger molecule in the nucleolar organizer. 5S rRNA is synthesized outside of nucleolus. The ribonucleoproteins are synthesized in the cytoplasm by usual mechanisms. It is in the nucleolus that newly synthesized rRNAs accumulate and become associated with required ribonucleoproteins and then migrate to the cytoplasm of cell in the form of ribosomal subunits. | |
==Function== | ==Function== | ||
| − | Ribosomes are the workhorses of [[protein biosynthesis]], the process of translating [[messenger RNA]] (mRNA) into [[protein]]. The mRNA comprises a series of [[codon]]s that dictate to the ribosome the sequence of the [[amino acid]]s needed to make the protein. Using the mRNA as a template, the ribosome traverses each codon of the mRNA, pairing it with the appropriate amino acid. This is done using molecules of [[transfer RNA]] (tRNA) containing a complementary [[anticodon]] on one end and the appropriate amino acid on the | + | Ribosomes are the workhorses of [[protein biosynthesis]], the process of translating [[messenger RNA]] (mRNA) into [[protein]]. The mRNA comprises a series of [[codon]]s that dictate to the ribosome the sequence of the [[amino acid]]s needed to make the protein. Using the mRNA as a template, the ribosome traverses each codon of the mRNA, pairing it with the appropriate amino acid. This is done using molecules of [[transfer RNA]] (tRNA) containing a complementary [[anticodon]] on one end and the appropriate amino acid on the another. |
| − | Protein synthesis begins at a [[start codon]] near the 5' end of the mRNA. The small ribosomal subunit, typically bound to a tRNA containing the amino acid [[methionine]], binds to an AUG codon on the mRNA and recruits the large ribosomal subunit. | + | Protein synthesis begins at a [[start codon]] near the 5' end of the mRNA. The small ribosomal subunit, typically bound to a tRNA containing the amino acid [[methionine]], binds to an AUG codon on the mRNA and recruits the large ribosomal subunit. Of the three tRNA binding sites, designated by A, P, and E, the A–site tRNA bears an incoming amino acid, and the P–site tRNA carries the growing peptide chain. Peptide bond formation attaches the peptide to the A–site tRNA's amino acid. The P–site tRNA then moves to the E–site (E stands for "exit"), replacing the former, uncharged E–site tRNA. The A–site tRNA, now bearing the growing peptide, is shifted into the P position. A new tRNA bearing the next amino acid is then brought into the A–site. |
| − | [[Image:ribosome.png|frame|none|'''Figure | + | [[Image:ribosome.png|frame|none|'''Figure 4 :''' Translation of mRNA (1) by a ribosome (2) into a <font color=#AA00AA>polypeptide chain</font> (3). The mRNA begins with a start [[codon]] (<font color="#00AA00">AUG</font>) and ends with a stop codon (<font color="#00AA00">UAG</font>).]] |
| − | In Figure | + | In Figure 4, both ribosomal subunits (<font color="#0000AA">small</font> and <font color="#AA0000">large</font>) assemble at the start codon (towards the 5' end of the mRNA). The ribosome uses [[tRNA]] that matches the current codon (triplet) on the mRNA to append an [[amino acid]] to the polypeptide chain. This is done for each triplet on the mRNA, while the ribosome moves toward the 3' end of the mRNA. Usually in bacterial cells, several ribosomes are working parallel on a single mRNA, forming what we call a ''polyribosome'' or ''[[polysome]]''. |
==References== | ==References== | ||
| − | + | *Alberts, B., A. Johnson, J. Lewis, M. Raff, K. Roberts, and P. Walter. 2002. ''The Molecular Biology of the Cell''. Garland Science. ISBN 0815332181 | |
| + | *Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. ''Science'' 289(5481): 905–920. | ||
| + | * Benne, R., and P. Sloof. 1987. [http://www.ncbi.nlm.nih.gov/pubmed/2446672 Evolution of the mitochondrial protein synthetic machinery]. ''BioSystems'' 21(1): 51–68. PMID 2446672. Retrieved July 17, 2008. | ||
| + | *O'Brien, T. W. 1971. The general occurrence of 55S ribosomes in mammalian liver mitochondria. ''J. Biol. Chem''. 245: 3409. | ||
| + | *Palade, G. E. 1955. A small particulate component of the cytoplasm. ''J. Biophys. Biochem. Cytol.'' 1(1): 59–68. | ||
| + | *Roberts, R. B. 1958. Introduction. In R. B. Roberts, ''Microsomal Particles and Protein Synthesis''. New York: Pergamon Press. | ||
| + | *Schluenzen F., A. Tocilj, R. Zarivach, J. Harms, M. Gluehmann, D. Janell, A. Bashan, H. Bartels, I. Agmon, F. Franceschi, and A. Yonath. 2000. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. ''Cell'' 102(5): 615–623. | ||
| + | *Schuwirth B. S., M. A. Borovinskaya, C. W. Hau, W. Zhang, A. Vila–Sanjurjo, J. M. Holton, and J. H. Cate. 2005. Structures of the bacterial ribosome at 3.5 Å resolution. ''Science'' 310(5749): 827–834. | ||
| + | *Selmer, M., C. M. Dunham, F. V. Murphy, A. Weixlbaumer, S. Petry, A. c. Kelley, J. R. Weir, and V. Ramakrishnan. 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. ''Science'' 313(5795): 1935–1942. | ||
| + | *Wimberly, B. T., D. E. Brodersen, W. M. Clemons, R. J. Morgan–Warren, A. P. Carter, C. Vonrhein, T. Hartsch, and V. Ramakrishnan. 2000. Structure of the 30S ribosomal subunit. ''Nature'' 407(6802): 327–339. | ||
| + | *Yusupov, M. M., G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. Cate, and H. F. Noller. 2001. Crystal structure of the ribosome at 5.5 Å resolution. ''Science'' 292(5518): 883–896. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | {{organelles}} | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | {{credit|113768938}} | + | {{credit|Ribosome|113768938|Transfer_RNA|152142549|Messenger_RNA|157780918|RNA_world_hypothesis|154232645}} |
[[Category:Life sciences]] | [[Category:Life sciences]] | ||
| + | [[Category:Cell biology]] | ||
Latest revision as of 09:20, 10 August 2022
A ribosome is a small, dense granular particle comprising usually three or four ribosomal RNA molecules and more than 50 protein molecules, interconnected to form the site of protein synthesis. The ribosome is the site at which the messenger RNA's code for linking amino acids together in a chain to form a particular new protein is translated into that protein or polypeptide. Ribosomes occur in both prokaryotic and eukaryotic cells. Ribosomes from bacteria and archaea are smaller than the ribosomes from eukaryotes, although all three domains of life have significantly different ribosomes. Interestingly, the ribosomes in the mitochondrion of eukaryotic cells resemble those in bacteria, reflecting the assumed evolutionary origin of this organelle (Benne and Sloof 1987).
A ribosome can be thought of as a giant enzyme that builds proteins from a set of genetic instructions. The ribosome's enzymatic activity derives from the presence of the rRNA, which in acting as an enzyme exemplifies a ribozyme and lends credence to the RNA world hypothesis that in the origins of life, RNA preceded DNA. Ribosomes occur either freely, as in the matrix of mitochondria, chloroplasts, and cytoplasm (the internal fluid of the cell), or in a membrane-bound state, as in the endoplasmic reticulum and the nuclear envelope.
The intricate process by which messenger RNA is translated into a specific sequence of amino acids is a testimony to the remarkable complexity and harmony in the universe. The process has to be very precise, otherwise the functionality of the protein could be compromised.
Overview
Ribosomes, the sites of protein synthesis within living cells, receive instructions from the DNA genes through messenger ribonucleic acid (mRNA), encoding a chemical "blueprint" for a protein product. A protein is a complex, high-molecular mass organic compound comprising amino acids joined together in chains, called peptides or polypeptides depending on their length. Transfer RNA (tRNA) is a small RNA chain (73-93 nucleotides) that transfers a specific amino acid to a growing polypeptide chain at the ribosomal site of protein synthesis during translation of the mRNA into a protein.
The rRNA portions of the ribosome perform the catalytic processes by which ribosomes synthesize proteins while the protein portions of the ribosome support the function of the rRNA ribozyme (an RNA molecule that catalyzes a chemical reaction). Such evidence lends support to the RNA world hypothesis, which proposes that a world filled with RNA (ribonucleic acid) based life predates current world filled with DNA (deoxyribonucleic acid) based life. In the proposed RNA world, RNA would have served some of the catalytic functions now served by proteins, and the ribosomes would be a remnant from that world now deposed by the DNA world.
Ribosomes were first clearly described by Romanian cell biologist George Palade in the mid–1950s, as dense particles or granules of ribonucleoprotein, after he observed them under the electron microscope (Palade 1955). For this, Palade would win the Nobel Prize. The term "ribosome" was later proposed by the scientist Richard B. Roberts in 1958, while writing the introductory comments for the symposium proceedings "Microsomal Particles and Protein Synthesis" (Roberts 1958).
The structure and function of the ribosomes and associated molecules, known as the translational apparatus, has been of research interest since the mid-20th century and the focus of the study has been to work out the topology (shape and positions of the individual protein and rRNA) of ribosomes.
Occurrence
Ribosomes are abundant components of both prokaryotic and eukaryotic cells and of both plant and animal cells. An Escherichia coli cell contains roughly 10,000 ribosomes, which together form about 25 percent of the total bacterial cell mass. A mammalian cell may contain as many as 10 million ribosomes. In prokaryotic cells, the ribosomes are distributed freely in the cytoplasm. In eukaryotic cells, they are found either freely floating in the matrix of mitochondria, chloroplasts, and cytoplasm or attached to the membrane of the endoplasmic reticulum and the nuclear envelope.
Free and membrane–bound ribosomes differ only in their spatial distribution; they are identical in structure and function. Whether the ribosome exists in a free or membrane–bound state depends on the presence of an ER targeting signal sequence on the protein being synthesized.
Free ribosomes
Free ribosomes are "free" to move about anywhere in the cytoplasm (within the cell membrane). Yeast cells, reticulocytes or lymphocytes, meristematic plant tissues, embryonic nerve cells, and cancerous cells contain a large number of free ribosomes. Proteins made by free ribosomes are used within the cell. Thus, the cells that synthesize specific proteins for the intracellular utilization and storage often contain large number of free ribosomes. Such cells include erythroblasts, developing muscle cells, skin cells, and so forth.
Membrane–bound ribosomes
When certain proteins are synthesized, they need be "membrane–bound." Therefore, the new polypeptide chains are usually synthesized in membrane-bound ribosomes and are inserted directly into the endoplasmic reticulum, from where they are then transported to their destinations. Bound ribosomes usually produce proteins that are used within the cell membrane or are expelled from the cell via exocytosis. Thus, in the cells actively engaged in protein synthesis, the ribosomes tend to remain attached to the membranes of the endoplasmic reticulum. Such cells include the pancreatic cells, hepatic parenchymal cells, osteoblasts, serous cells, or submaxillary gland cells, mammary gland cells, thyroid cells, and the chief cells of the glandular stomach in birds, rodents, and ruminants.
Structure
Overview
The various ribosomes share a core structure that is quite similar despite the large differences in size. Ribosomes are oblate spheroid granules with a diameter ranging from 15 to 25 nanometers (150 to 250 Å). Each ribosome is porous, hydrated, and consists of two subunits (Figure 1). One ribosomal subunit is larger in size and has a dome–like shape, while the other ribosomal subunit is smaller and occurs above the larger one forming a cap–like structure (Figure 2). The ribosomes are chemically composed mainly of RNA (ribosomal RNA, rRNA) and proteins and thus are called ribonucleoproteins, RNPs. Both constituents occur approximately in equal proportion in its two subunits. Lipid is totally absent in ribosomes.
Crystallographic work has shown that there are no ribosomal proteins close to the reaction site for polypeptide synthesis, which is fully occupied by the rRNA. This suggests that the ribosomal protein does not participate directly in the synthesis of proteins, but rather provides a scaffold that enhances the ability of rRNA to synthesize protein (Alberts et al. 2002).
The two ribosomal subunits remain fit together due to a high concentration of Mg++ ions. In a decreased Mg++ concentration, the two subunits dissociate. Actually, in bacterial cells, the two subunits are found to occur freely in the cytoplasm and they come together only for the process of protein synthesis. At high concentration of Mg++ ions in the matrix, two ribosomes (each called monosomes) become associated with each other and form what is known as dimer. Further, during the process of protein synthesis, several ribosomes are often working their way along the same mRNA, somewhat like beads on a string; the resulting structure is known as polyribosome or polysome.
Prokaryotic ribosomes
Prokaryotes have comparatively smaller ribosomes with the sedimentation coefficient of 70 Svedberg units (abbreviated as S), and a molecular weight of 2.7x106 daltons. Each of the 70S ribosomes comprises a small (30S) and a large (50S) subunit. (Svedberg measures are not additive because sedimentation rate depends on both mass and surface area.) The 70S ribosomes contain proportionally more RNA than protein. For example, the ribosomes of E. coli contain 63 percent rRNA and 37 percent protein. The 70S ribosomes have three different types of rRNA: 23S rRNA, 16S rRNA, and 5S rRNA. The large subunit is composed of a 5S rRNA subunit (comprising 120 nucleotides), a 23S rRNA subunit (comprising 2900 nucleotides), and 34 proteins. The 30S subunit has a 16S rRNA subunit (comprising 1540 nucleotides) bound to 21 proteins (Alberts et al. 2002).
Eukaryotic ribosomes
Eukaryotes have bigger ribosomes of 80S sedimentation coefficient and of 40x106 daltons molecular weight. Each 80S ribosome comprises a small (40S) and a large (60S) subunit. The ribosomal subunits of prokaryotes and eukaryotes are quite similar (Alberts et al. 2002). However, 80S ribosomes are composed of proportionally less RNA and more protein. For example, in pea seedling, ribosomes comprise 40 percent rRNA and 60 percent protein.
There are four different types of rRNA in 80S ribosomes: 28S rRNA (but 25–26S rRNA in plants, fungi, and protozoans), 18S rRNA, 5S rRNA, and 5.8S rRNA. The large 60S subunit comprises a 5S RNA (120 nucleotides), a 28S RNA (4700 nucleotides), and a 5.8S RNA (160 nucleotides) subunit, and about 49 proteins. The 40S subunit comprises an 18S RNA (1900 nucleotides) subunit and about 33 proteins (Alberts et al. 2002). About 60 percent of the rRNA is helical (i.e., double stranded) and contains paired bases. These double stranded regions are due to hairpin loops between complimentary regions of the linear molecule. Thus, it can be said that the extra RNA in the larger ribosomes is in several long continuous insertions, such that they form loops out of the core structure without disrupting or changing it (Alberts et al. 2002).
The ribosomes found in chloroplasts and mitochondria of eukaryotes also comprise large and small subunits bound together into one 55S particle (Alberts et al. 2002). These organelles are believed to be descendants of bacteria (see Endosymbiotic theory) and as such their ribosomes are similar to those of prokaryotes (Alberts et al. 2002). The 55S ribosomes of mammalian mitochondria lack 5S rRNA, but contain 21S and 12S rRNAs. The 21S rRNA occurs in larger or 35S ribosomal subunit, while 12S rRNA occurs in smaller or 25S ribosomal subunit.
The differences between the prokaryotic and eukaryotic ribosomes are exploited by pharmaceutical chemists to create antibiotics that can destroy a bacterial infection without harming the cells of the infected person. Due to differences in their structures, the bacterial 70S ribosomes are vulnerable to these antibiotics (such as Chloramphenicol) while the eukaryotic 80S ribosomes are not. Even though mitochondria possess ribosomes similar to the bacterial ones, mitochondria are not affected by these antibiotics because they are surrounded by a double membrane that does not easily admit these antibiotics into the organelle (O'Brien 1971).
Ultra–structure
The general molecular structure of the ribosome has been known since the early 1970s. In the early 2000s, the structure was revealed at high resolutions, in the order of a few angstroms, and scientific papers presenting the structure of the ribosome at atomic resolution were published in rapid succession in late 2000.
The structure of the 30S small subunit from Thermus thermophilus (a highly thermophilic bacteria first discovered in deep–sea hot vents) shows that the decoding center, which positions mRNA and three tRNAs, is constructed entirely of RNA (Schluenzen et al., 2000; Wimberly, 2000). The mRNA threads through a tunnel within the small subunit. The 3’ end of the 16S rRNA is supposed to be involved in mRNA binding. Each of the three tRNAs is bound in distinctive binding sites made from structural elements contributed by both the 50S subunit and the 30S subunit. In each of the three tRNA binding sites (A–, P–, and E–sites), the ribosome contacts all of the major elements of tRNA, providing an explanation for the conservation of tRNA structure (Yusupov et al. 2001). The anticodon stem–loops of tRNAs point into the 30S subunit, whereas 3’ ends attached to amino acid or peptide through an acyl bond point down in 50S subunit. Further, the 3’ ends of the A–site and P–site tRNAs are juxtaposed in the peptidyl transferase site of the 50S subunit. However, a metal ion stabilizes a kink in the mRNA that demarcates the boundary between A and P sites, which is potentially important to prevent slippage of mRNA, while translocation of tRNA from site A to P. Also, metal ions stabilize the inter–subunit interface (Selmer 2006).
The 16S rRNA of the small, 30S subunit folds into four domains: 5', central, 3' major, and 3' minor. The structural autonomy of these domains implies that they move relative to one another. Thus, the head of the small subunit shows a high degree of flexibility compared to its rest of the body. Swiveling of the head observed by Schuwirth et al. (2005) suggests a mechanism for the final movements of messenger RNA (mRNA) and transfer RNAs (tRNAs) during translocation. Structural changes correlating events at the particle's far end with the cycle of mRNA translocation at the decoding region are transmitted by extended RNA helical elements that run longitudinally through its body (Schluenzen et al. 2000).
The 23S rRNA of the large, 50S subunit folds into six secondary structural domains, while the seventh domain is formed by 5S rRNA. The 50S subunit from the archea, Haloarcula marismortui is shown to have all its rRNAs fit together like the pieces of a three–dimensional jigsaw puzzle to form a large, monolithic structure. Proteins are found everywhere on its surface except in the active site where peptide bond formation takes place and where it contacts the small subunit. Most of the proteins stabilize the structure by interacting with several rRNA domains (Ban et al. 2000). The large ribosomal subunit catalyzes peptide bond formation and binds factors for initiation, termination, and elongation. The peptidyl transferase function is attributed to the 23S rRNA, making this RNA a "ribozyme." Nascent polypeptides emerge through a tunnel in the large ribosome subunit. The tunnel lumen is lined with RNA helices and some ribosomal protein.
Biogenesis
Ribosomes are not self–replicating particles. Synthesis of various components of ribosomes such as rRNAs and proteins is under genetic control. In bacteria, a single gene transcript containing the sequences for 16S, 23S, and 5S rRNAs is synthesized by an rRNA operon (transcriptional unit for multiple molecules) and this larger molecule undergoes both tailoring and chemical modifications before each rRNA molecule assumes its mature form. The whole process of biosynthesis of 70S ribosomes takes place in the cytoplasm. The required amount of ribonucleoprotein synthesis is under autogenous regulation of translation.
In eukaryotes, the biogenesis of ribosomes is much more complex involving three main events: rRNA synthesis by nucleolar organizer (NO), synthesis of 5S rRNA, and biosynthesis of ribonucleoprotein. The 5.8S, 18S, and 28S rRNAs are transcribed as a much larger molecule in the nucleolar organizer. 5S rRNA is synthesized outside of nucleolus. The ribonucleoproteins are synthesized in the cytoplasm by usual mechanisms. It is in the nucleolus that newly synthesized rRNAs accumulate and become associated with required ribonucleoproteins and then migrate to the cytoplasm of cell in the form of ribosomal subunits.
Function
Ribosomes are the workhorses of protein biosynthesis, the process of translating messenger RNA (mRNA) into protein. The mRNA comprises a series of codons that dictate to the ribosome the sequence of the amino acids needed to make the protein. Using the mRNA as a template, the ribosome traverses each codon of the mRNA, pairing it with the appropriate amino acid. This is done using molecules of transfer RNA (tRNA) containing a complementary anticodon on one end and the appropriate amino acid on the another.
Protein synthesis begins at a start codon near the 5' end of the mRNA. The small ribosomal subunit, typically bound to a tRNA containing the amino acid methionine, binds to an AUG codon on the mRNA and recruits the large ribosomal subunit. Of the three tRNA binding sites, designated by A, P, and E, the A–site tRNA bears an incoming amino acid, and the P–site tRNA carries the growing peptide chain. Peptide bond formation attaches the peptide to the A–site tRNA's amino acid. The P–site tRNA then moves to the E–site (E stands for "exit"), replacing the former, uncharged E–site tRNA. The A–site tRNA, now bearing the growing peptide, is shifted into the P position. A new tRNA bearing the next amino acid is then brought into the A–site.
In Figure 4, both ribosomal subunits (small and large) assemble at the start codon (towards the 5' end of the mRNA). The ribosome uses tRNA that matches the current codon (triplet) on the mRNA to append an amino acid to the polypeptide chain. This is done for each triplet on the mRNA, while the ribosome moves toward the 3' end of the mRNA. Usually in bacterial cells, several ribosomes are working parallel on a single mRNA, forming what we call a polyribosome or polysome.
ReferencesISBN links support NWE through referral fees
- Alberts, B., A. Johnson, J. Lewis, M. Raff, K. Roberts, and P. Walter. 2002. The Molecular Biology of the Cell. Garland Science. ISBN 0815332181
- Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289(5481): 905–920.
- Benne, R., and P. Sloof. 1987. Evolution of the mitochondrial protein synthetic machinery. BioSystems 21(1): 51–68. PMID 2446672. Retrieved July 17, 2008.
- O'Brien, T. W. 1971. The general occurrence of 55S ribosomes in mammalian liver mitochondria. J. Biol. Chem. 245: 3409.
- Palade, G. E. 1955. A small particulate component of the cytoplasm. J. Biophys. Biochem. Cytol. 1(1): 59–68.
- Roberts, R. B. 1958. Introduction. In R. B. Roberts, Microsomal Particles and Protein Synthesis. New York: Pergamon Press.
- Schluenzen F., A. Tocilj, R. Zarivach, J. Harms, M. Gluehmann, D. Janell, A. Bashan, H. Bartels, I. Agmon, F. Franceschi, and A. Yonath. 2000. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell 102(5): 615–623.
- Schuwirth B. S., M. A. Borovinskaya, C. W. Hau, W. Zhang, A. Vila–Sanjurjo, J. M. Holton, and J. H. Cate. 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310(5749): 827–834.
- Selmer, M., C. M. Dunham, F. V. Murphy, A. Weixlbaumer, S. Petry, A. c. Kelley, J. R. Weir, and V. Ramakrishnan. 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313(5795): 1935–1942.
- Wimberly, B. T., D. E. Brodersen, W. M. Clemons, R. J. Morgan–Warren, A. P. Carter, C. Vonrhein, T. Hartsch, and V. Ramakrishnan. 2000. Structure of the 30S ribosomal subunit. Nature 407(6802): 327–339.
- Yusupov, M. M., G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. Cate, and H. F. Noller. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292(5518): 883–896.
| Organelles of the cell |
|---|
| Acrosome | Chloroplast | Cilium/Flagellum | Centriole | Endoplasmic reticulum | Golgi apparatus | Lysosome | Melanosome | Mitochondrion | Myofibril | Nucleus | Parenthesome | Peroxisome | Plastid | Ribosome | Vacuole | Vesicle |
Credits
New World Encyclopedia writers and editors rewrote and completed the Wikipedia article in accordance with New World Encyclopedia standards. This article abides by terms of the Creative Commons CC-by-sa 3.0 License (CC-by-sa), which may be used and disseminated with proper attribution. Credit is due under the terms of this license that can reference both the New World Encyclopedia contributors and the selfless volunteer contributors of the Wikimedia Foundation. To cite this article click here for a list of acceptable citing formats.The history of earlier contributions by wikipedians is accessible to researchers here:
The history of this article since it was imported to New World Encyclopedia:
Note: Some restrictions may apply to use of individual images which are separately licensed.