Ribozyme
Ribozyme (from ribonucleic acid enzyme) is a ribonucleic acid (RNA) molecule that can catalyze biochemical reactions, just as as certain protein enzymes act as biological catalysts in regulating the rate of chemical reactions in living organisms. These RNA enzymes have been found in various organelles (nucleus, mitochondrion, chloroplast) in eukaryote organisms, as well as in some viruses, including bacterial viruses (Robinson 2003). Ribozymes can either be involved in cleaving RNA chains or binding RNA molecules together, and appear also to be involved in the linkage of amino acids into proteins at the ribosome (Robinson 2003). A ribozyme also is known as catalytic RNA.
Unraveling the mystery of the ribozyme not only has scientific interest, but also potential medical application. Ribozymes can be developed to cleave particular sequences of RNAs, which, if incorporated by human cell, could make dysfunctional particular RNA viruses.
Overview and function
An enzyme is a biological catalyst that regulates the rate of a chemical reaction in a living organism. Many biochemical reactions, such as the cleavage of a molecule or the binding together of two molecules, requires catalysts (Robinson 2003). These catalysts accelerate the rate of the chemical reactions. Most enzymes are proteins, and for a long time it was assumed that proteins alone had sufficient structural complexity to serve as catalysts in living organisms (Robinson 2003). However, around 1980 it was discovered that certain nucleic acids, called ribozymes, are also capable of catalytic activity (Robinson 2003).
There are different classes of ribozymes, but all appear to be associated with metal ions, such as potassium and magnesium (Robinson 2003). Different ribozymes catalyze different reactions, but almost all ribozymes are involved in catalyzing the cleavage of RNA chains or the formation of bonds between RNA strands (Robinson 2003). Many natural ribozymes catalyze either the hydrolysis of one of their own phosphodiester bonds, or the hydrolysis of bonds in other RNAs. However, RNA is not the only target, as ribozymes also have been found to catalyze the aminotransferase activity of the ribosome.
Although most ribozymes are quite rare in the cell, their roles are sometimes essential to life. For example, the functional part of the ribosome, the molecular machine that translates RNA into proteins, is fundamentally a ribozyme.
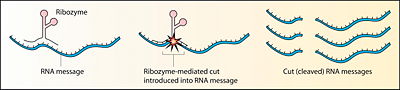
Many ribozymes have either a hairpin- or hammerhead-shaped active center and a unique secondary structure that allows them to cleave RNA molecules at specific sequences (Kanchana 2007). It is now possible to make ribozymes that will specifically cleave any RNA molecule. These RNA catalysts may have medical applications. For example, a ribozyme has been designed to cleave the RNA of HIV. If such a ribozyme was made by a cell, all incoming virus particles would have their RNA genome cleaved by the ribozyme, which would prevent infection (Kanchana 2007).
Some ribozymes may play an important role as therapeutic agents, as enzymes which tailor defined RNA sequences, as biosensors, and for applications in functional genomics and gene discovery (Hean and Weinberg 2008).
A recent test-tube study of prion folding suggests that an RNA may catalyze the pathological protein conformation in the manner of a chaperone enzyme (Supattapone 2004).
Known ribozymes
Naturally occurring ribozymes include:
- Peptidyl transferase 23S rRNA
- RNase P
- Group I and Group II introns
- GIR1 branching ribozyme (Nielsen et al. 2005)
- Leadzyme‚ÄĒAlthough initially created in vitro, natural examples have been found
- Hairpin ribozyme
- Hammerhead ribozyme
- HDV ribozyme
- Mammalian CPEB3 ribozyme
- VS ribozyme
- glmS ribozyme
- CoTC ribozyme
Discovery
Before the discovery of ribozymes, protein enzymes were the only known biological catalysts. In 1967, Carl Woese, Francis Crick, and Leslie Orgel suggested that RNA also could act as a catalyst. This idea was based upon the discovery that RNA can form complex secondary structures (Woese 1967). The term ribozyme was first introduced by Kelly Kruger et al. in 1982, in a paper published in Cell.
The first ribozymes were reported in the 1980s by Thomas R. Cech, who was studying RNA splicing in the ciliated protozoan Tetrahymena thermophila, and Sidney Altman, who was working on the bacterial RNase-P complex. These ribozymes were found in the intron of an RNA transcript, which removed itself from the transcript, as well as in the RNA component of the RNase-P complex, which is involved in the maturation of pre-tRNAs.
Essentially, Cech was studying the excision of introns in a ribosomal RNA gene in Tetrahymena thermophila. While trying to purify the enzyme responsible for the splicing, he found that the intron could be spliced out even in the absence of any added cell extract. Cech and his colleagues could not identify any protein associated with the splicing reaction, and proposed that the intron sequence portion of the RNA itself could break and reform phosphodiester bonds (Kanchana 2007).
At about the same time, Altman was studying the way tRNA molecules are processed in the cell when he and his colleagues isolated an enzyme called Ribonuclease P (RNase P), whose function is to cleave off an extra, or precursor, sequence of RNA on tRNA molecules, resulting in conversion of a precursor tRNA into the active tRNA. To their surprise, Altman and his colleagues found that RNase P contained RNA in addition to protein and that RNA was an essential component of the active enzyme. This finding was so foreign to the current paradigm that they had difficulty publishing their findings. Subsequently, Altman demonstrated that RNA can act as a catalyst by showing that the RNase P RNA submit could catalyze the cleavage of precursor tRNA into active tRNA in the absence of any protein component (Kanchana 2007).
In 1989, Thomas R. Cech and Sidney Altman won the Nobel Prize in Chemistry for their "discovery of catalytic properties of RNA" (NP 1989).
Since Cech's and Altman's discovery, other investigators have discovered other examples of self-cleaving RNAs or catalytic RNA molecules (Kanchana 2007).
Ribozymes and "RNA world"
At one point, it was held that catalysis was reserved for proteins. In retrospect, the occurrence of catalytic RNA makes a lot of sense, for it addresses an old question regarding the origin of life: "Which came first, enzymes that do the work of the cell or nucleic acids that carry the information required to produce the enzymes" (Kanchana 2007). Having nucleic acids also serve as catalysts circumvents this problem (Kanchana 2007; Visser 1984).
RNA can act as a hereditary molecule, which encouraged Walter Gilbert to propose that in the past, the cell used RNA as both the genetic material and the structural and catalytic molecule, rather than dividing these functions between DNA and protein as they are today. This hypothesis became known as the "RNA world hypothesis" of the origin of life.
If ribozymes were the first molecular machines used by early life, then today's remaining ribozymes‚ÄĒsuch as the ribosome machinery‚ÄĒcould be considered living fossils of a life based primarily on nucleic acids.
Investigators studying the origin of life have produced ribozymes in the laboratory that are capable of catalyzing their own synthesis under very specific conditions, such as an RNA polymerase ribozyme (Johnston et al. 2001). Mutagenesis and selection has been performed resulting in isolation of improved variants of the "Round-18" polymerase ribozyme from 2001. "B6.61" is able to add up to 20 nucleotides to a primer template in 24 hours, until it decomposes by hydrolysis of its phosphodiester bonds (Zaher and Unrau 2007).
Artificial ribozymes
Since the discovery of ribozymes that exist in living organisms, there has been interest in the study of new synthetic ribozymes made in the laboratory. For example, artificially-produced self-cleaving RNAs that have good enzymatic activity have been produced. Tang and Breaker (1997) isolated self-cleaving RNAs by in vitro selection of RNAs originating from random-sequence RNAs. Some of the synthetic ribozymes that were produced had novel structures, while some were similar to the naturally occurring hammerhead ribozyme.
The techniques used to discover artificial ribozymes involve using selection pressures. This approach takes advantage of RNA's dual nature as both a catalyst and an informational polymer, making it easy for an investigator to produce vast populations of RNA catalysts using polymerase enzymes. The ribozymes are mutated by reverse transcribing them with reverse transcriptase into various cDNA and amplified with mutagenic PCR. The selection parameters in these experiments often differ. One approach for selecting a ligase ribozyme involves using biotin tags, which are covalently linked to the substrate. If a molecule possesses the desired ligase activity, a streptavidin matrix can be used to recover the active molecules.
ReferencesISBN links support NWE through referral fees
- Doherty, E.A., and J.A. Doudna. 2001. Ribozyme structures and mechanisms. Annu Rev Biophys Biomol Struct 30:457-475. Retrieved July 11, 2008.
- Hean, J., and M.S. Weinberg. 2008. The hammerhead ribozyme revisited: New biological insights for the development of therapeutic agents and for reverse genomics applications. In K.V. Morris, RNA and the Regulation of Gene Expression: A Hidden Layer of Complexity. Caister Academic Press. ISBN 9781904455257.
- Ikawa, Y., K. Tsuda, S. Matsumura, and T. Inoue. De novo synthesis and development of an RNA enzyme. PNAS 101(38): 13750-13755. Retrieved July 11, 2008.
- Johnston, W., P. Unrau, M. Lawrence, M. Glasner, and D. Bartel. 2001. RNA-catalyzed RNA polymerization: Accurate and general RNA-templated primer extension. Science 292(5520): 1319‚Äď1325. Retrieved July 11, 2008.
- Joyce, G.G. 2004. Directed evolution of nucleic acid enzymes. Annu Rev Biochem 73:791-836. Retrieved July 11, 2008.
- Kanchana, P. 2007. Not all enzymatic reactions are catalyzed by protein. Pulashti.info
- Kruger, K., P.J. Grabowski, A.J. Zaug, J. Sands, D.E. Gottschling, and T.R. Cech. 1982. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of tetrahymena. Cell 31: 147-157.
- Nielsen, H., E. Westhof, and S. Johansen. 2005. An mRNA is capped by a 2', 5' lariat catalyzed by a group I-like ribozyme. Science 309(5740): 1584‚Äď1587. Retrieved July 11, 2008.
- Nobel Prize. 1989. The Nobel Prize in Chemistry 1989 "for their discovery of catalytic properties of RNA": Sidney Altman and Thomas R. Cech. NobelPrize.org. Retrieved July 11, 2008.
- Robinson, R. 2003. Genetics. New York: Macmillan Reference USA. ISBN 0028658906.
- Supattapone, S. 2004. Prion protein conversion in vitro. Journal of Molecular Medicine 82: 348-356.
- Tang, J., and R. R. Breaker. 1997. Structural diversity of self-cleaving ribozymes. Proceedings of the National Academy of Sciences 97(11): 5784‚Äď5789. Retrieved July 11, 2008.
- Visser, C.M. 1984. Evolution of biocatalysis. Part one. Possible pre-genetic RNA catalysts which are their own replicas. Origins of Life 14(1-4): 291-300. Retrieved July 11, 2008.
- Woese, C. 1967. The Genetic Code. New York: Harper and Row.
- Zaher, H. S., and P. Unrau. 2007. Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA 13(7): 1017‚Äď26. Retrieved July 11, 2008.
Credits
New World Encyclopedia writers and editors rewrote and completed the Wikipedia article in accordance with New World Encyclopedia standards. This article abides by terms of the Creative Commons CC-by-sa 3.0 License (CC-by-sa), which may be used and disseminated with proper attribution. Credit is due under the terms of this license that can reference both the New World Encyclopedia contributors and the selfless volunteer contributors of the Wikimedia Foundation. To cite this article click here for a list of acceptable citing formats.The history of earlier contributions by wikipedians is accessible to researchers here:
The history of this article since it was imported to New World Encyclopedia:
Note: Some restrictions may apply to use of individual images which are separately licensed.