Difference between revisions of "Messenger RNA" - New World Encyclopedia
Rick Swarts (talk | contribs) (added article from Wikipedia and credit/category tag) |
Rick Swarts (talk | contribs) |
||
| Line 2: | Line 2: | ||
The "life cycle" of an '''mRNA''' in a eukaryotic cell. RNA is [[transcription (genetics)|transcribed]] in the [[cell nucleus|nucleus]]; once completely processed, it is transported to the [[cytoplasm]] and [[Translation (genetics)|translated]] by the [[ribosome]]. At the end of its life, the mRNA is degraded.]] | The "life cycle" of an '''mRNA''' in a eukaryotic cell. RNA is [[transcription (genetics)|transcribed]] in the [[cell nucleus|nucleus]]; once completely processed, it is transported to the [[cytoplasm]] and [[Translation (genetics)|translated]] by the [[ribosome]]. At the end of its life, the mRNA is degraded.]] | ||
| − | '''Messenger ribonucleic acid''' ('''mRNA''') is a | + | '''Messenger ribonucleic acid''' ('''mRNA''') is a class of [[ribonucleic acid]] (RNA) molecules that serve as chemical "blueprints" for the production of a [[protein]] product, carrying the coding information from a [[DNA]] template to the [[ribosome]]s, where the transcription into proteins takes place. |
| + | |||
| + | Messenger RNA is synthesized on a DNA template in a process known as DNA [[transcription (genetics)|transcription]]. In mRNA, as in DNA, genetic information is encoded in the sequence of four [[nucleotides]] arranged into [[codon]]s of three bases each. Each codon encodes for a specific [[amino acid]], except the [[stop codon]]s that terminate protein synthesis. The mRNAs then carry this information for protein synthesis to the sites of protein synthesis (ribosomes). Here, transfer RNAs (tRNAs) bind on one end to specific codons (three-base region) in the mRNA and bind on the other end to the amino acids specified by that codon, and thus places the amino acids in the correct sequence in the growing polypeptide according to the template (sequence of nucleotides) provided by the mRNA (Alberts et al. 1989). That is, the nucleic acid polymer is [[translation (genetics)|translated]] into a protein. This process requires two other types of RNA: [[transfer RNA]] (tRNA), which mediates recognition of the codon and provides the corresponding amino acid, and [[ribosomal RNA]] (rRNA), which is the central component of the ribosome's protein manufacturing machinery. | ||
==Processing and function== | ==Processing and function== | ||
Revision as of 19:51, 6 October 2008
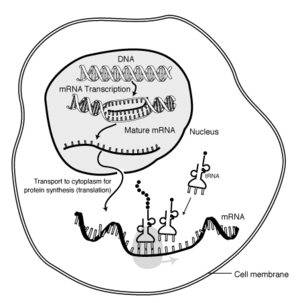
Messenger ribonucleic acid (mRNA) is a class of ribonucleic acid (RNA) molecules that serve as chemical "blueprints" for the production of a protein product, carrying the coding information from a DNA template to the ribosomes, where the transcription into proteins takes place.
Messenger RNA is synthesized on a DNA template in a process known as DNA transcription. In mRNA, as in DNA, genetic information is encoded in the sequence of four nucleotides arranged into codons of three bases each. Each codon encodes for a specific amino acid, except the stop codons that terminate protein synthesis. The mRNAs then carry this information for protein synthesis to the sites of protein synthesis (ribosomes). Here, transfer RNAs (tRNAs) bind on one end to specific codons (three-base region) in the mRNA and bind on the other end to the amino acids specified by that codon, and thus places the amino acids in the correct sequence in the growing polypeptide according to the template (sequence of nucleotides) provided by the mRNA (Alberts et al. 1989). That is, the nucleic acid polymer is translated into a protein. This process requires two other types of RNA: transfer RNA (tRNA), which mediates recognition of the codon and provides the corresponding amino acid, and ribosomal RNA (rRNA), which is the central component of the ribosome's protein manufacturing machinery.
Processing and function
The brief existence of an mRNA molecule begins with transcription and ultimately ends in degradation. During its life, an mRNA molecule may also be processed, edited, and transported prior to translation. Eukaryotic mRNA molecules often require extensive processing and transport, while prokaryotic molecules do not.
Transcription
During transcription, RNA polymerase makes a copy of a gene from the DNA to mRNA as needed. This process is similar in eukaryotes and prokaryotes. One notable difference, however, is that eukaryotic RNA polymerase associates with mRNA processing enzymes during transcription so that processing can proceed quickly after the start of transcription. The short-lived, unprocessed or partially processed, product is termed pre-mRNA; once completely processed, it is termed mature mRNA.
Eukaryotic pre-mRNA processing
Processing of mRNA differs greatly among eukaryotes, bacteria and archea. Non-eukaryotic mRNA is essentially mature upon transcription and requires no processing, except in rare cases. Eukaryotic pre-mRNA, however, requires extensive processing.
5' cap addition
A 5' cap (also termed an RNA cap, an RNA 7-methylguanosine cap or an RNA m7G cap) is a modified guanine nucleotide that has been added to the "front" or 5' end of a eukaryotic messenger RNA shortly after the start of transcription. The 5' cap consists of a terminal 7-methylguanosine residue which is linked through a 5'-5'-triphosphate bond to the first transcribed nucleotide. Its presence is critical for recognition by the ribosome and protection from RNases.
Cap addition is coupled to transcription, and occurs co-transcriptionally, such that each influences the other. Shortly after the start of transcription, the 5' end of the mRNA being synthesized is bound by a cap-synthesizing complex associated with RNA polymerase. This enzymatic complex catalyzes the chemical reactions that are required for mRNA capping. Synthesis proceeds as a multi-step biochemical reaction.
Splicing
Splicing is the process by which pre-mRNA is modified to remove certain stretches of non-coding sequences called introns; the stretches that remain include protein-coding sequences and are called exons. Sometimes pre-mRNA messages may be spliced in several different ways, allowing a single gene to encode multiple proteins. This process is called alternative splicing. Splicing is usually performed by an RNA-protein complex called the spliceosome, but some RNA molecules are also capable of catalyzing their own splicing (see ribozymes).
Editing
In some instances, an mRNA will be edited, changing the nucleotide composition of that mRNA. An example in humans is the apolipoprotein B mRNA, which is edited in some tissues, but not others. The editing creates an early stop codon, which upon translation, produces a shorter protein.
Polyadenylation
Polyadenylation is the covalent linkage of a polyadenylyl moiety to a messenger RNA molecule. In eukaryotic organisms, most messenger RNA (mRNA) molecules are polyadenylated at the 3' end. The poly(A) tail and the protein bound to it aid in protecting mRNA from degradation by exonucleases. Polyadenylation is also important for transcription termination, export of the mRNA from the nucleus, and translation. mRNA can also be polyadenylated in prokaryotic organisms, where poly(A) tails act to facilitate, rather than impede, exonucleolytic degradation.
Polyadenylation occurs during and immediately after transcription of DNA into RNA. After transcription has been terminated, the mRNA chain is cleaved through the action of an endonuclease complex associated with RNA polymerase. After the mRNA has been cleaved, around 250 adenosine residues are added to the free 3' end at the cleavage site. This reaction is catalyzed by polyadenylate polymerase. Just as in alternative splicing, there can be more than one polyadenylation variant of a mRNA.
Transport
Another difference between eukaryotes and prokaryotes is mRNA transport. Because eukaryotic transcription and translation is compartmentally separated, eukaryotic mRNAs must be exported from the nucleus to the cytoplasm. Mature mRNAs are recognized by their processed modifications and then exported through the nuclear pore.
Translation
Because prokaryotic mRNA does not need to be processed or transported, translation by the ribosome can begin immediately after the end of transcription. Therefore, it can be said that prokaryotic translation is coupled to transcription and occurs co-transcriptionally.
Eukaryotic mRNA that has been processed and transported to the cytoplasm (i.e. mature mRNA) can then be translated by the ribosome. Translation may occur at ribosomes free-floating in the cytoplasm, or directed to the endoplasmic reticulum by the signal recognition particle. Therefore, unlike prokaryotes, eukaryotic translation is not directly coupled to transcription.
Degradation
After a certain amount of time, the message is degraded by RNases. The limited lifetime of mRNA enables a cell to alter protein synthesis rapidly in response to its changing needs.
Different mRNAs within the same cell have distinct lifetimes (stabilities). In bacterial cells, individual mRNAs can survive from seconds to more than an hour; in mammalian cells, mRNA lifetimes range from several minutes to days. The greater the stability of an mRNA, the more protein may be produced from that mRNA. The presence of AU-rich elements in some mammalian mRNAs tends to destabilize those transcripts through the action of cellular proteins that bind these motifs. Rapid mRNA degradation via AU-rich elements is a critical mechanism for preventing the overproduction of potent cytokines such as tumor necrosis factor (TNF) and granulocyte-macrophage colony stimulating factor (GM-CSF).[1] Base pairing with a small interfering RNA (siRNA) or microRNA (miRNA) can also accelerate mRNA degradation.
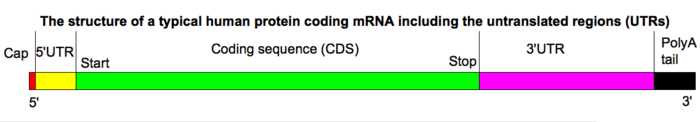
Structure
5' cap
The 5' cap is a modified guanine nucleotide added to the "front" (5' end) of the pre-mRNA using a 5',5-Triphosphate linkage. This modification is critical for recognition and proper attachment of mRNA to the ribosome, as well as protection from 5' exonucleases. It may also be important for other essential processes, such as splicing and transport.
Coding regions
Coding regions are composed of codons, which are decoded and translated into one (mostly eukaryotes) or several (mostly prokaryotes) proteins by the ribosome. Coding regions begin with the start codon and end with the a stop codons. Generally, the start codon is an AUG triplet and the stop codon is UAA, UAG, or UGA. The coding regions tend to be stabilised by internal base pairs, this impedes degradation.[2][3] In addition to being protein-coding, portions of coding regions may serve as regulatory sequences in the pre-mRNA as exonic splicing enhancers or exonic splicing silencers.
Untranslated regions
Untranslated regions (UTRs) are sections of the mRNA before the start codon and after the stop codon that are not translated, termed the five prime untranslated region (5' UTR) and three prime untranslated region (3' UTR), respectively. These regions are transcribed with the coding region and thus are exonic as they are present in the mature mRNA. Several roles in gene expression have been attributed to the untranslated regions, including mRNA stability, mRNA localization, and translational efficiency. The ability of a UTR to perform these functions depends on the sequence of the UTR and can differ between mRNAs.
The stability of mRNAs may be controlled by the 5' UTR and/or 3' UTR due to varying affinity for RNA degrading enzymes called ribonucleases and for ancillary proteins that can promote or inhibit RNA degradation.
Translational efficiency, including sometimes the complete inhibition of translation, can be controlled by UTRs. Proteins that bind to either the 3' or 5' UTR may affect translation by influencing the ribosome's ability to bind to the mRNA. MicroRNAs bound to the 3' UTR also may affect translational efficiency or mRNA stability.
Cytoplasmic localization of mRNA is thought to be a function of the 3' UTR. Proteins that are needed in a particular region of the cell can actually be translated there; in such a case, the 3' UTR may contain sequences that allow the transcript to be localized to this region for translation.
Some of the elements contained in untranslated regions form a characteristic secondary structure when transcribed into RNA. These structural mRNA elements are involved in regulating the mRNA. Some, such as the SECIS element, are targets for proteins to bind. One class of mRNA element, the riboswitches, directly bind small molecules, changing their fold to modify levels of transcription or translation. In these cases, the mRNA regulates itself.
Poly(A) tail
The 3' poly(A) tail is a long sequence of adenine nucleotides (often several hundred) at the 3' end of the pre-mRNA. This tail promotes export from the nucleus and translation, and protects the mRNA from degradation.
Monocistronic versus polycistronic mRNA
An mRNA molecule is said to be monocistronic when it contains the genetic information to translate only a single protein. This is the case for most of the eukaryotic mRNAs[4]. On the other hand, polycistronic mRNA carries the information of several genes, which are translated into several proteins. These proteins usually have a related function and are grouped and regulated together in an operon. Most of the mRNA found in bacteria and archea are polycistronic[4]. Dicistronic is the term used to describe a mRNA that encodes only two proteins.
ReferencesISBN links support NWE through referral fees
- ↑ Shaw G, Kamen R (August 1986). A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46 (5): 659–67.
- ↑ Shabalina SA, Ogurtsov AY, Spiridonov NA (2006). A periodic pattern of mRNA secondary structure created by the genetic code. Nucleic Acids Res. 34 (8): 2428–37.
- ↑ Katz L, Burge CB (September 2003). Widespread selection for local RNA secondary structure in coding regions of bacterial genes. Genome Res. 13 (9): 2042–51.
- ↑ 4.0 4.1 Kozak, M. (March 1983). Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiological Reviews 47 (1): 1–45.
External links
- Life of mRNA Flash animation
| Nucleic acids edit |
|---|
| Nucleobases: Adenine - Thymine - Uracil - Guanine - Cytosine - Purine - Pyrimidine |
| Nucleosides: Adenosine - Uridine - Guanosine - Cytidine - Deoxyadenosine - Thymidine - Deoxyguanosine - Deoxycytidine |
| Nucleotides: AMP - UMP - GMP - CMP - ADP - UDP - GDP - CDP - ATP - UTP - GTP - CTP - cAMP - cGMP |
| Deoxynucleotides: dAMP - dTMP - dUMP - dGMP - dCMP - dADP - dTDP - dUDP - dGDP - dCDP - dATP - dTTP - dUTP - dGTP - dCTP |
| Nucleic acids: DNA - RNA - LNA - PNA - mRNA - ncRNA - miRNA - rRNA - siRNA - tRNA - mtDNA - Oligonucleotide |
Credits
New World Encyclopedia writers and editors rewrote and completed the Wikipedia article in accordance with New World Encyclopedia standards. This article abides by terms of the Creative Commons CC-by-sa 3.0 License (CC-by-sa), which may be used and disseminated with proper attribution. Credit is due under the terms of this license that can reference both the New World Encyclopedia contributors and the selfless volunteer contributors of the Wikimedia Foundation. To cite this article click here for a list of acceptable citing formats.The history of earlier contributions by wikipedians is accessible to researchers here:
The history of this article since it was imported to New World Encyclopedia:
Note: Some restrictions may apply to use of individual images which are separately licensed.