Genetic recombination
Genetic recombination is an aspect of the variation that is observed in the genetic inheritance as it is passed down the generations by sexual reproduction. It underlys the changes that occur during evolution but it is not well understood. Much work is needed before we understand this aspect of God's creative process.
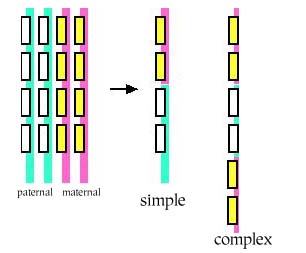
The genetic information is carried on the chromosomes, one set inhereted from the father, one set inherited from the mother. When a pair of chromosomes 'recobine' during sex cell formation, in the simplest case genetic information on the paternal chromosome is switched with information on the maternal on, and vice versa. The two 'daughter' chromosomes that result are not identical to either parental ones.
The diploid germ cell which divides into the sex cells had two copies of the genome (each as a double helix so there are four strands of DNA involved). The egg and sperm haploid sex cells that result have one copy of the genome (one set of chromosomes as a single double helix). The process of genetic recombination (or crossing ove') occurs at the stage of this meiosis sex-cell creation when there are four copies of the genome present (involving eight strands of DNA) all alligned with one another on the spindle axis.
This state where eight copies of DNA are being mixed and matched only last about 15 mins. It invloves many other molecules (RNA and proteins) and the complex is called the tetraplex or tetrad state. Guided by this complex of RNA and proteins, a variable amount of cutting and pasting between these eight strands occurs. The genetic material is organized in a modular fashion, and the shuffling involves these modules. It is currently an open question as to whether this tetraplex reorganization of the genetic inheritance is random (as classical science would have it, or is as well organized and well-designed as is the ribosome-triplet code method of protein synthesis.
A well-characterized example of such DNA manipulation by RNA/protein complexes is to be found in the make-antibody programming of an immature lymphocyte in the immune system. This is directed variation within a well-defined permutation space. The antibody-program carried by the immature lympocyte is then run in a virtual-body environment generated by the thymus. If the program running in this virtual realityrecognizes a body component, the lymphocycte is destroyed, else it is activated and released as a mature T-cell.
This immune system manipulation of DNA modules has been a focus of study as it has great relevance to disease. The DNA manipulation in the tetraplex stage, on the other hand, is not well-characterized, but the basic principles are probably similar.
The simplest type of recombination is crossing over, where strands are cut across and then reconnected differently. Even this apsect is highly modular; there are hot spots where crossing over occurs frequently, and forbidden zones where it never happens.
The most complex rearrangement of the genetic modules occurs during speciation. For instance, along the lineage that diverged from the apes tohumans, two chromosomes became fused into one (the human Chromosome Two, the second largest one). Such massive changes and manipulation of the genome by the RNA-protein complex are not well understood.
Genetic recombination is, more technically put, the transmission-genetic process by which the combinations of alleles observed at different loci (plural of locus) in two parental individuals become shuffled in offspring individuals. This definition is commonly used in classical transmission genetics, evolutionary biology, and population genetics. Such shuffling can be the result of recombination via intra-chromososomal recombination (crossing over) and via inter-chromososomal recombination (also called independent assortment). In evolutionary biology, genetic recombination, be it inter- or intra-chromososomal, is thought to have many advantages including that of allowing sexually reproducing organisms to avoid Muller's ratchet.
In molecular biology, recombination generally refers to the molecular process by which genetic variation found associated at two different places in a continuous piece of DNA becomes disassociated (shuffled). In this process one or both of the genetic variants are replaced by different variants found at same two places in a second DNA molecule. One mechanism leading to such molecular recombination is chromosomal crossing over. Such shuffling of variation is also possible between duplicated loci within the same DNA molecule. If the number of loci in each of the recombinant molecules is changed by the shuffling process, one speaks of "unbalanced" recombination or unequal crossing over. Enzymes called recombinases catalyze this reaction.
Crossing over
Main article: Chromosomal crossover
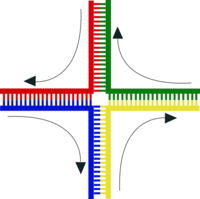
Crossing over of two chromosomes inherited from one's parents occurs during meiosis. After chromosomal replication, the four available chromatids are in tight formation with one another. During this time, homologous sites on two chromatids can mesh with one another, and may exchange genetic information. Immediately after replication, the tetrad formed by replication contains two pairs of two identical chromatids; after crossing over, each of the four chromatids carries a unique set of genetic information.
Chemistry of crossover
Enzymes known as recombinases catalyze the reactions that allow for crossover to occur. A recombinase creates a nick in one strand of a DNA double helix, allowing the nicked strand to pull apart from its complementary strand and anneal to one strand of the double helix on the opposite chromatid. A second nick allows the unannealed strand in the second double helix to pull apart and anneal to the remaining strand in the first, forming a structure known as a cross-strand exchange or a Holliday junction. The Holliday junction is a tetrahedral structure which can be 'pulled' by other recombinases, moving it along the four-stranded structure.
Consequences of crossover
In most eukaryotes, a cell carries two copies of each gene, each referred to as an allele. Each parent passes on one allele to each offspring. Even without recombination, each gamete contains a random assortment of chromatids, choosing randomly from each pair of chromatids available. With recombination, however, the gamete can receive a (mostly) random assortment of individual genes, as each chromosome may contain genetic information from two different chromatids.
Recombination results in a new arrangement of maternal and paternal alleles on the same chromosome. Although the same genes appear in the same order, the alleles are different. This process explains why offspring from the same parents can look so different. In this way, it is theoretically possible to have any combination of parental alleles in an offspring, and the fact that two alleles appear together in one offspring does not have any influence on the statistical probability that another offspring will have the same combination. This theory of "independent assortment" of alleles is fundamental to genetic inheritance. However, there is an exception that requires further discussion.
The frequency of recombination is actually not the same for all gene combinations. This is because recombination is greatly influenced by the proximity of one gene to another. If two genes are located close together on a chromosome, the likelihood that a recombination event will separate these two genes is less than if they were farther apart. Genetic linkage describes the tendency of genes to be inherited together as a result of their location on the same chromosome. Linkage disequilibrium describes a situation in which some combinations of genes or genetic markers occur more or less frequently in a population than would be expected from their distances apart. This concept is applied when searching for a gene that may cause a particular disease. This is done by comparing the occurrence of a specific DNA sequence with the appearance of a disease. When a high correlation between the two is found, it is likely that the appropriate gene sequence is closer.
Problems of crossover
Crossover recombination can occur between any two double helices of DNA which are very close in sequence and come into contact with one another. Thus, crossover may occur between Alu repeats on the same chromatid, or between similar sequences on two completely different chromosomes. These processes are called unbalanced recombination. Unbalanced recombination is fairly rare compared to normal recombination, but severe problems can arise if a gamete containing unbalanced recombinants becomes part of a zygote. Offspring with severe unbalances rarely live through birth.
Other types of recombination
Conservative site-specific recombination
In conservative site-specific recombination, a mobile DNA element is inserted into a strand of DNA by means similar to that seen in crossover. A segment of DNA on the mobile element matches exactly with a segment of DNA on the target, allowing enzymes called integrases to insert the rest of the mobile element into the target.
Transpositional recombination
Another form of site-specific recombination, transpositional recombination does not require an identical strand of DNA in the mobile element to match with the target DNA. Instead, the integrases involved introduce nicks in both the mobile element and the target DNA, allowing the mobile DNA to enter the sequence. The nicks are then removed by ligases.
Illegitimate recombination
Recombination between DNA sequences that contain only a few identical nucleotides.
External links
ReferencesISBN links support NWE through referral fees
- Alberts, B. et al., Molecular Biology of the Cell, 3rd Edition. Garland Publishing, 1994.
- Mayerhofer R, Koncz-Kalman Z, Nawrath C, Bakkeren G, Crameri A, Angelis K, Redei GP, Schell J, Hohn B, Koncz C. T-DNA integration: a mode of illegitimate recombination in plants. EMBO J. 1991 Mar;10(3):697-704.
- This article contains material from the Science Primer published by the NCBI, which, as a US government publication, is in the public domain at http://www.ncbi.nlm.nih.gov/About/disclaimer.html.
Credits
New World Encyclopedia writers and editors rewrote and completed the Wikipedia article in accordance with New World Encyclopedia standards. This article abides by terms of the Creative Commons CC-by-sa 3.0 License (CC-by-sa), which may be used and disseminated with proper attribution. Credit is due under the terms of this license that can reference both the New World Encyclopedia contributors and the selfless volunteer contributors of the Wikimedia Foundation. To cite this article click here for a list of acceptable citing formats.The history of earlier contributions by wikipedians is accessible to researchers here:
The history of this article since it was imported to New World Encyclopedia:
Note: Some restrictions may apply to use of individual images which are separately licensed.