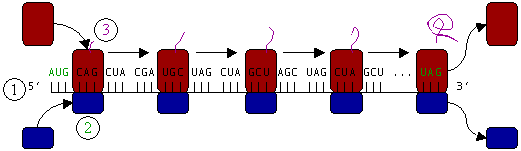Ribosome
A ribosome is a small, dense granular particle of ribonucleoprotein found in both prokaryotic as well as eukaryotic cells. It is the protein synthesizing machinary that translates messenger RNA (mRNA) into a polypeptide chain (e.g., a protein) of amino acids delivered by Transfer RNA (tRNA). It can be thought of as a giant enzyme that builds a protein from a set of genetic instructions. Ribosomes occur either freely in the matrix of mitochondria, chloroplanst and cytoplasm (the internal fluid of the cell) or in membrane bound state in the endoplasmic reticulum and the nuclear envelope. Since ribosomes are ribozymes, it is thought that they might be remnants of the RNA world.
Ribosomes were first clearly described by Romanian cell biologist George Palade in the mid-1950s as dense particles or granules of ribonucleoprotein after observing under the electron microscope[1] for which he would win the Nobel Prize. The term ribosome was later proposed by the scientist Richard B. Roberts in 1958 while writing the introductory comments:
During the course of the symposium a semantic difficulty became apparent. To some of the participants, microsomes mean the ribonucleoprotein particles of the microsome fraction contaminated by other protein and lipid material; to others, the microsomes consist of protein and lipid contaminated by particles. The phrase “microsomal particles” does not seem adequate, and “ribonucleoprotein particles of the microsome fraction” is much too awkward. During the meeting the word “ribosome” was suggested; this seems a very satisfactory name, and it has a pleasant sound. The present confusion would be eliminated if “ribosome” were adopted to designate ribonucleoprotein particles in the size range 20 to 100S.
Roberts, R. B., Microsomal Particles and Protein Synthesis[2]
The structure and function of the ribosomes and associated molecules, known as the translational apparatus, has been of research interest since the mid 20th century and the focus of the study has been to work out the topology (shape and positions of the individual protein and rRNA) of ribosomes.
(1) Head. (2) Platform. (3) Base. (4) Ridge. (5) Central protuberance. (6) Back. (7) Stalk. (8) Front.
Occurrence
The ribosomes occur both in prokaryotic as well as eukaryotic cells and both in plant as well as animal cell. An E. coli cell contains 10,000 ribosomes, forming about 25 percent of the total bacterial cell mass. A cultured mammalian cell may contain as much as 10 million ribosomes. In prokaryotic cells, the ribosomes are distributed freely in the cytoplasm. In eukaryotic cells, they are found either freely foating in the matrix of mitochondria, chloroplanst and cytoplasm or attached to the membrane of the endoplasmic reticulum and the nuclear envelope. Free and membrane-bound ribosomes differ only in their spatial distribution; they are identical in structure and function. Whether the ribosome exists in a free or membrane-bound state depends on the presence of a ER-targeting signal sequence on the protein being synthesized.
Free ribosomes
Free ribosomes are "free" to move about anywhere in the cytoplasm (within the cell membrane). The yeast cells, reticulocytes or lymphocytes, meristematic plant tissues, embryonic nerve cells and cancerous cells contain large number of free ribosomes. Proteins made by free ribosomes are used within the cell. Thus, the cells which synthesize specific proteins for the intracellular utilization and storage often contain large number of free ribosomes. Such cells are the erythroblasts, developing muscle cells, skin cells, etc.
Membrane-bound ribosomes
When certain proteins are synthesized, they need be "membrane-bound". Therefore, the new polypeptide chains are usually synthesized in membrane bound ribosomes and are inserted directly into the endoplasmic reticulum, from where they are then transported to their destinations. Bound ribosomes usually produce proteins that are used within the cell membrane or are expelled from the cell via exocytosis. Thus, in the cells actively engagged in protein synthesis, the ribosomes remain attached to the membranes of the endoplasmic reticulum. Such cells are the pancreatic cells, plasma cells, hepatic parenchymal cells, Nissls bodies, osteoblasts, serous cells or submaxillary gland cells, chief cells of the glandular stomach, thyroid cells and mammary gland cells.
Structure
Overview
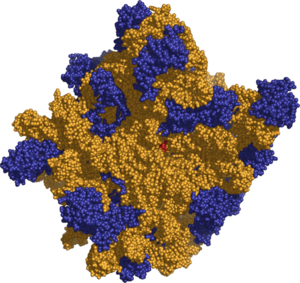
The various ribosomes share a core structure which is quite similar despite the large differences in size. Ribosomes are oblate spheroid granules with a diameter ranging from 15 to 25nm (150 to 250 A). Each ribosome is porous, hydrated and consists of two subunits (Figure 1). One ribosomal subunit is larger in size and has a dome-like shape, while the other ribosomal subunit is smaller and occurs above the larger one forming a cap-like structure (Figure 2). The ribosomes are chemically composed of mainly RNA (ribosomal RNA, rRNA) and proteins (ribonucleoprotein, RNP). Lipid is totally absent in ribosomes. Both constituents occur approximately in equal proportion in its two subunits.
Crystallographic work has shown that there are no ribosomal proteins close to the reaction site for polypeptide synthesis. This suggests that the protein components of ribosomes act as a scaffold that may enhance the ability of rRNA to synthesize protein rather than directly participating in catalysis. Also it has been pointed out that all of the catalytic activity of the ribosome is carried out by the RNA, the proteins reside on the surface and seem to stabilize the structure.[3] (See: Ribozyme). The two ribosomal subunits remain fitted together due to high concentration of Mg++ ions. In the decreased Mg++ concentration, the two subunits dissociate. Actually, in bacterial cells, the two subunits are found to occur freely in the cytoplasm and they come together only for the process of protein synthesis. At high concentration of Mg++ ions in the matrix, two ribosomes (each called monosomes) become associated with each other and form what is known as dimer. Further, during the process of protein synthesis, many ribosomes are woven together just like beads due to a common mRNA; the resulting structure is known as polyribosome or polysome.
Prokaryotic Ribosomes
Prokaryotes have comparatively smaller ribosomes with the sedimentation coefficient of 70 Svedberg unit (abbreviated as S), and a molecular weight of 2.7x106 daltons. Each of the 70S ribosomes consists of a small (30S) and a large (50S) subunits. The 70S ribosomes contain proportionally more RNA than protein. For example, the ribosomes of E. coli contain 63 percent rRNA and 37 percent protein. The 70S ribosomes have three different types of rRNA, viz., 23S rRNA, 16S rRNA and 5S rRNA. The large subunit is composed of a 5S rRNA subunit (consisting of 120 nucleotides), a 23S rRNA subunit (with 2900 nucleotides) and 34 proteins. The 30S subunit has a 16S rRNA subunit (with 1540 nucleotides) bound to 21 proteins.[3]
Eukaryotic Ribosomes
Eukaryotes have bigger ribosomes of 80S sedimentation coefficient and of 40x106 daltons molecular weight. Each 80S ribosome consists of a small (40S) and large (60S) subunits. The ribosomal subunits of prokaryotes and eukaryotes are quite similar.[3] However, 80S ribosomes are composed of proportionally less RNA and more protein. For example, in pea seedling, ribosomes have 40 percent rRNA and 60 percent protein. There are four different types of rRNA in 80S ribosomes, viz., 28S rRNA (but 25-26S rRNA in plants, fungi and protozoans), 18S rRNA, 5S rRNA and 5.8S rRNA. The large 60S subunit is composed of a 5S RNA (120 nucleotides), a 28S RNA (4700 nucleotides), a 5.8S RNA (160 nucleotides) subunits and ~49 proteins. The 40S subunit has a 18S RNA (1900 nucleotides) subunit and ~33 proteins.[3] About 60 percent of the rRNA is helical (i.e., double stranded) and contains paired bases. These double standed regions are due to hairpin loops between complimentary regions of the linear molecule. Thus we can say that the extra RNA in the larger ribosomes is in several long continuous insertions, such that they form loops out of the core structure without disrupting or changing it[3].
The ribosomes found in chloroplasts and mitochondria of eukaryotes also consist of large and small subunits bound together into one 55S particle[3]. These organelles are believed to be descendants of bacteria (see Endosymbiotic theory) and as such their ribosomes are similar to those of prokaryotes.[4] The 55S ribosomes of mammalian mitochondria lack 5S rRNA, but contain 21S and 12S rRNAs. The 21S rRNA occurs in larger or 35S ribosomal subunit, while 12S rRNA occur in smaller or 25S ribosomal subunit.
The differences between the prokaryotic and eukaryotic ribosomes are exploited by pharmaceutical chemists to create antibiotics that can destroy a bacterial infection without harming the cells of the infected person. Due to the differences in their structures, the bacterial 70S ribosomes are vulnerable to these antibiotics (such as Chloramphenicol) while the eukaryotic 80S ribosomes are not. Even though mitochondria possess ribosomes similar to the bacterial ones, mitochondria are not affected by these antibiotics because they are surrounded by a double membrane that does not easily admit these antibiotics into the organelle[5].
Ultrastructure
The general molecular structure of the ribosome has been known since the early 1970s. In the early 2000s the structure has been achieved at high resolutions, in the order of a few angstroms.
The first papers giving the structure of the ribosome at atomic resolution, were published in rapid succession in late 2000. First, the 50S (large prokaryotic) subunit from the archea, Haloarcula marismortui was published[6]. Soon after the structure of the 30S subunit from Thermus thermophilus was published.[7] Shortly thereafter a more detailed structure was published.[8] Early the next year (May 2001) these coordinates were used to reconstruct the entire T. thermophilus 70S particle at 5.5 Å resolution[9].
Two papers were published in November 2005 with structures of the Escherichia coli 70S ribosome. The structures of vacant ribosome were determined at 3.5 Å resolution using x-ray crystallography [10]. Then two weeks later a structure based on cryo-electron microsopy was published[11] which depicts the ribosome at 11-15 Å in the act of passing a newly synthesized protein strand into the protein-conducting channel.
First atomic structures of the ribosome complexed with tRNA and mRNA molecules were solved by using X-ray crystallography by two groups independently, at 2.8 Å [12] and at 3.7 Å [13]. These structures allow to see the details of interactions of the Thermus thermophilus ribosome with mRNA and with tRNAs bound at classical ribosomal sites. Interactions of the ribosome with long mRNAs containing Shine-Dalgarno sequences were visualized soon after that at 4.5 to 5.5-Å resolution [14].
Function
Ribosomes are the workhorses of protein biosynthesis, the process of translating messenger RNA (mRNA) into protein. The mRNA comprises a series of codons that dictate to the ribosome the sequence of the amino acids needed to make the protein. Using the mRNA as a template, the ribosome traverses each codon of the mRNA, pairing it with the appropriate amino acid. This is done using molecules of transfer RNA (tRNA) containing a complementary anticodon on one end and the appropriate amino acid on the other.
Protein synthesis begins at a start codon near the 5' end of the mRNA. The small ribosomal subunit, typically bound to a tRNA containing the amino acid methionine, binds to an AUG codon on the mRNA and recruits the large ribosomal subunit. The large ribosomal subunit contains three tRNA binding sites, designated A, P, and E. The A site binds an aminoacyl-tRNA (a tRNA bound to an amino acid); the P site binds a peptidyl-tRNA (a tRNA bound to the peptide being synthesized); and the E site binds a free tRNA before it exits the ribosome.
In Figure 3, both ribosomal subunits (small and large) assemble at the start codon (towards the 5' end of the mRNA). The ribosome uses tRNA which matches the current codon (triplet) on the mRNA to append an amino acid to the polypeptide chain. This is done for each triplet on the mRNA, while the ribosome moves towards the 3' end of the mRNA. Usually in bacterial cells, several ribosomes are working parallel on a single mRNA, forming what we call a polyribosome or polysome.
ReferencesISBN links support NWE through referral fees
- ↑ G.E. Palade. (1955) "A small particulate component of the cytoplasm." J Biophys Biochem Cytol. Jan;1(1): pages 59-68. PMID 14381428
- ↑ Roberts, R. B., editor. (1958) "Introduction" in Microsomal Particles and Protein Synthesis. New York: Pergamon Press, Inc.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 The Molecular Biology of the Cell, fourth eddition. Brusce Alberts, et al. Garland Science (2002) pg. 342 ISBN 0-8153-3218-1
- ↑ The Molecular Biology of the Cell, fourth edition. Brusce Alberts, et al. Garland Science (2002) pg. 808 ISBN 0-8153-3218-1
- ↑ O'Brien, T.W., The General Occurrence of 55S Ribosomes in Mammalian Liver Mitochondria. J. Biol. Chem., 245:3409 (1971).
- ↑ Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000 Aug 11;289(5481):905-20. PMID 10937989
- ↑ Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000 Sep 1;102(5):615-23. PMID 11007480
- ↑ Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000 Sep 21;407(6802):327-39. PMID 11014182
- ↑ Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001 May 4;292(5518):883-96. Epub 2001 Mar 29. PMID 11283358
- ↑ Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005 Nov 4;310(5749):827-34. PMID 16272117
- ↑ Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks CL 3rd, Ban N, Frank J. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature. 2005 Nov 17;438(7066):318-24. PMID 16292303
- ↑ Selmer, M., Dunham, C.M., Murphy, F.V IV, Weixlbaumer, A., Petry S., Kelley, A.C., Weir, J.R. and Ramakrishnan, V. (2006). Structure of the 70S ribosome complexed with mRNA and tRNA. Science , 313, 1935-1942
- ↑ Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006 Sep 22;126(6):1065-77
- ↑ Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006 Nov 16;444(7117):391-4
See also
|
|
External links
- 70S Ribosome Architecture Animation of a working ribosome. Requires the Chime browser plugin from this site (where registration is required).
- Lab computer simulates ribosome in motion
- Information on ribosomes
- Molecule of the Month © RCSB Protein Data Bank:
| Organelles of the cell |
|---|
| Acrosome | Chloroplast | Cilium/Flagellum | Centriole | Endoplasmic reticulum | Golgi apparatus | Lysosome | Melanosome | Mitochondrion | Myofibril | Nucleus | Parenthesome | Peroxisome | Plastid | Ribosome | Vacuole | Vesicle |
This article contains material from the Science Primer published by the NCBI, which, as a US government publication, is in the public domain at http://www.ncbi.nlm.nih.gov/About/disclaimer.html.
Credits
New World Encyclopedia writers and editors rewrote and completed the Wikipedia article in accordance with New World Encyclopedia standards. This article abides by terms of the Creative Commons CC-by-sa 3.0 License (CC-by-sa), which may be used and disseminated with proper attribution. Credit is due under the terms of this license that can reference both the New World Encyclopedia contributors and the selfless volunteer contributors of the Wikimedia Foundation. To cite this article click here for a list of acceptable citing formats.The history of earlier contributions by wikipedians is accessible to researchers here:
The history of this article since it was imported to New World Encyclopedia:
Note: Some restrictions may apply to use of individual images which are separately licensed.