Trichoplax
| Trichoplax adhaerens | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | ||||||||||||||
| Scientific classification | ||||||||||||||
| ||||||||||||||
| Trichoplax adhaerens F.E. von Schultze, 1883 |
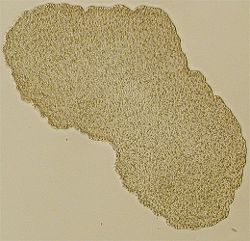
Trichoplax adhaerens is a simple balloon-like marine animal with a body cavity filled with pressurized fluid. It is given its own phylum, called Placozoa, sometimes referred to commonly as the tablet animals, which is a direct translation from the Greek scientific name; the only other species assigned to this taxon, Treptoplax reptans, was described in 1896 and has not been seen since, leading to doubts about its existence.
Individual Trichoplax are soft-bodied, about 0.5 mm across, and somewhat resemble a large amoeba. The name T. adhaerens was given because it tends to stick to its substrate, including glass pipettes and microscope slides. Its evolutionary relationships are still being investigated, but it may be allied with the cnidarians and ctenophores. Dellaporta et al have reported the complete mitochondrial genome sequence of Trichoplax adhaerens and showed that Placozoa are the most basal (that is, they branched off earliest) living eumetazoan phylum.[1]
Trichoplax lacks organs and most tissues, including nerve cells and a nervous system, although evidence suggests that they evolved from species with nerve cells.[citation needed] It is made up of a few thousand cells of four types in three distinct layers: monociliated dorsal and ventral epithelia cells, ventral gland cells and the syncytial fiber cells. But sensory cells and muscle cells are apparently absent. The outermost layer (the monociliated cells) have a single cilium, which allow the adult to move. The epithelia of Trichoplax lack a basal membrane and the cells are connected by belt desmosomes. Lipid inclusions, called 'shiny spheres', are regularly distributed over the dorsal epithelia.
It feeds by absorption and has been observed to form temporary bulges to trap food. It climbs atop its food and uses the ventral surface as a temporary extraorganismal gastric cavity. Digestion is both extracellular and by phagocytosis.
When not feeding Trichoplax is actively motile with movement effected by ventral ciliation and by the fiber cell layer and lacks any polarity in its movement.
Mansi Srivastava and her colleagues drew the first genome draft for Trichoplax in 2008[2]. They estimated that there are about 11,514 protein coding genes in Trichoplax genome. The genome size for Trichoplax is about 98 million base pair.
The haploid number of chromosomes is six. It has the smallest amount of DNA yet measured for any animal with only 50 megabases (80 femtograms per cell). A Trichoplax genome sequencing project has recently been completed.[3]
Putative eggs have been observed, but they degrade at the 32-64 cell stage. Neither embryonic development nor sperm have been observed, however Trichoplax genomes show evidence of sexual reproduction.[4] Asexual reproduction by binary fission is the primary mode of reproduction observed in the lab.
Trichoplax were discovered on the walls of a marine aquarium in the 1880s, and have rarely been observed in their natural habitat.[5] The full extent of their natural range is unknown, but they are easily collected in tropical and subtropical latitudes around the world.
Of the 11,514 genes identified in the six chromosomes of Trichoplax, 80% are shared with cnidarians and bilaterians. Trichoplax also shares over 80% of its introns—the regions within genes that are not translated into proteins—with humans. This junk DNA forms the basis of regulatory gene networks. The arrangement of genes is conserved between the Trichoplax and human genomes. This contrasts to other model systems such as fruit flies and soil nematodes that have experienced a paring down of non-coding regions and a loss of the ancestral genome organizations. [6]
Significant genetic differences have been observed between collected specimens matching the morphological description of T. adhaerens, suggesting that it may be a cryptic species complex.[7]
ReferencesISBN links support NWE through referral fees
- ↑ Dellaporta et al (2006). 'Mitochondrial genome of Trichoplax adhaerens supports Placozoa as the basal lower metazoan phylum'. Proceedings of the National Academy of Sciences 103 (23): 8751–6.
- ↑ Mansi Srivastava, Emina Begovic, Jarrod Chapman, Nicholas H. Putnam, Uffe Hellsten, Takeshi Kawashima, Alan Kuo, Therese Mitros, Asaf Salamov, Meredith L. Carpenter, Ana Y. Signorovitch, Maria A. Moreno, Kai Kamm, Jane Grimwood, Jeremy Schmutz, Harris Shapiro, Igor V. Grigoriev, Leo W. Buss, Bernd Schierwater, Stephen L. Dellaporta & Daniel S. Rokhsar (21 August 2008). The Trichoplax genome and the nature of placozoans. Nature 454 (7207): 955-960.
- ↑ Trichoplax Genome Sequenced - Rosetta Stone for Understanding Evolution.
- ↑ Signorovitch AY, Dellaporta SL, Buss LW (2005). Molecular signatures for sex in the Placozoa. Proceedings of the National Academy of Sciences 102 (43): 15518–22.
- ↑ Maruyama YK (2004). Occurrence in the field of a long-term, year-round, stable population of placozoans. Biol Bull 206 (1): 55–60.
- ↑ Primitive Pancake at Phyorg.com, Based on a DOE/Joint Genome Institute news release. Aug 30, 2008
- ↑ Voigt, O and Collins AG, Pearse VB, Pearse JS, Hadrys H, Ender A (2004). Placozoa — no longer a phylum of one. Current Biology 14 (22): R944–5.
Howey, R. L. http://www.microscopy-uk.org.uk/mag/indexmag.html?http://www.microscopy-uk.org.uk/mag/artoct98/tricho.html
http://www.ucmp.berkeley.edu/phyla/placozoa/placozoa.html
Introduction to Placozoa
The Most Simple of All Known Animals
Allen G. Collins,
University of California Museum of Paleontology
External links
- Movies of Trichoplax
- The Trichoplax Genome Project at the Yale Peabody Museum
- A Weird Wee Beastie: Trichoplax adhaerens
- Research articles from the ITZ, TiHo Hannover
- Information page from the University of California at Berkeley
- Ender A, Schierwater B (January 2003). Placozoa are not derived cnidarians: evidence from molecular morphology. Mol. Biol. Evol. 20 (1): 130–4. - Mitochondrial DNA and 16S rRNA analysis and phylogeny of Trichoplax adhaerens
- Historical overview of Trichoplax research
- Science Daily:Genome Of Simplest Animal Reveals Ancient Lineage, Confounding Array Of Complex Capabilities
Credits
New World Encyclopedia writers and editors rewrote and completed the Wikipedia article in accordance with New World Encyclopedia standards. This article abides by terms of the Creative Commons CC-by-sa 3.0 License (CC-by-sa), which may be used and disseminated with proper attribution. Credit is due under the terms of this license that can reference both the New World Encyclopedia contributors and the selfless volunteer contributors of the Wikimedia Foundation. To cite this article click here for a list of acceptable citing formats.The history of earlier contributions by wikipedians is accessible to researchers here:
The history of this article since it was imported to New World Encyclopedia:
Note: Some restrictions may apply to use of individual images which are separately licensed.